Worldwide Protein Data Bank validation information: usage and trends
- PMID: 29533231
- PMCID: PMC5947764
- DOI: 10.1107/S2059798318003303
Worldwide Protein Data Bank validation information: usage and trends
Abstract
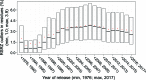
Realising the importance of assessing the quality of the biomolecular structures deposited in the Protein Data Bank (PDB), the Worldwide Protein Data Bank (wwPDB) partners established Validation Task Forces to obtain advice on the methods and standards to be used to validate structures determined by X-ray crystallography, nuclear magnetic resonance spectroscopy and three-dimensional electron cryo-microscopy. The resulting wwPDB validation pipeline is an integral part of the wwPDB OneDep deposition, biocuration and validation system. The wwPDB Validation Service webserver (https://validate.wwpdb.org) can be used to perform checks prior to deposition. Here, it is shown how validation metrics can be combined to produce an overall score that allows the ranking of macromolecular structures and domains in search results. The ValTrendsDB database provides users with a convenient way to access and analyse validation information and other properties of X-ray crystal structures in the PDB, including investigating trends in and correlations between different structure properties and validation metrics.
Keywords: PDB; Protein Data Bank; X-ray crystallography; quality control; three-dimensional macromolecular structure; validation.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

