Polyamine Deacetylase Structure and Catalysis: Prokaryotic Acetylpolyamine Amidohydrolase and Eukaryotic HDAC10
- PMID: 29533602
- PMCID: PMC5988950
- DOI: 10.1021/acs.biochem.8b00079
Polyamine Deacetylase Structure and Catalysis: Prokaryotic Acetylpolyamine Amidohydrolase and Eukaryotic HDAC10
Abstract
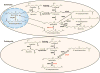
Polyamines such as putrescine, spermidine, and spermine are small aliphatic cations that serve myriad biological functions in all forms of life. While polyamine biosynthesis and cellular trafficking pathways are generally well-defined, only recently has the molecular basis of reversible polyamine acetylation been established. In particular, enzymes that catalyze polyamine deacetylation reactions have been identified and structurally characterized: histone deacetylase 10 (HDAC10) from Homo sapiens and Danio rerio (zebrafish) is a highly specific N8-acetylspermidine deacetylase, and its prokaryotic counterpart, acetylpolyamine amidohydrolase (APAH) from Mycoplana ramosa, is a broad-specificity polyamine deacetylase. Similar to the greater family of HDACs, which mainly serve as lysine deacetylases, both enzymes adopt the characteristic arginase-deacetylase fold and employ a Zn2+-activated water molecule for catalysis. In contrast with HDACs, however, the active sites of HDAC10 and APAH are sterically constricted to enforce specificity for long, slender polyamine substrates and exclude bulky peptides and proteins containing acetyl-l-lysine. Crystal structures of APAH and D. rerio HDAC10 reveal that quaternary structure, i.e., dimer assembly, provides the steric constriction that directs the polyamine substrate specificity of APAH, whereas tertiary structure, a unique 310 helix defined by the P(E,A)CE motif, provides the steric constriction that directs the polyamine substrate specificity of HDAC10. Given the recent identification of HDAC10 and spermidine as mediators of autophagy, HDAC10 is rapidly emerging as a biomarker and target for the design of isozyme-selective inhibitors that will suppress autophagic responses to cancer chemotherapy, thereby rendering cancer cells more susceptible to cytotoxic drugs.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

