Bcr-Abl regulation of sphingomyelin synthase 1 reveals a novel oncogenic-driven mechanism of protein up-regulation
- PMID: 29533737
- PMCID: PMC6044059
- DOI: 10.1096/fj.201701016R
Bcr-Abl regulation of sphingomyelin synthase 1 reveals a novel oncogenic-driven mechanism of protein up-regulation
Abstract
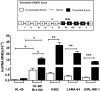
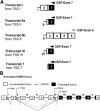
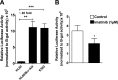
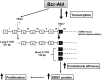
Bcr-Abl (break-point cluster region-abelson), the oncogenic trigger of chronic myelogenous leukemia (CML), has previously been shown to up-regulate the expression and activity of sphingomyelin synthase 1 (SMS1), which contributes to the proliferation of CML cells; however, the mechanism by which this increased expression of SMS1 is mediated remains unknown. In the current study, we show that Bcr-Abl enhances the expression of SMS1 via a 30-fold up-regulation of its transcription. Of most interest, the Bcr-Abl-regulated transcription of SMS1 is initiated from a novel transcription start site (TSS) that is just upstream of the open reading frame. This shift in TSS utilization generates an SMS1 mRNA with a substantially shorter 5' UTR compared with its canonical mRNA. This shorter 5' UTR imparts a 20-fold greater translational efficiency to SMS1 mRNA, which further contributes to the increase of its expression in CML cells. Therefore, our study demonstrates that Bcr-Abl increases SMS1 protein levels via 2 concerted mechanisms: up-regulation of transcription and enhanced translation as a result of the shift in TSS utilization. Remarkably, this is the first time that an oncogene-Bcr-Abl-has been demonstrated to drive such a mechanism that up-regulates the expression of a functionally important target gene, SMS1.-Moorthi, S., Burns, T. A., Yu, G.-Q., Luberto, C. Bcr-Abl regulation of sphingomyelin synthase 1 reveals a novel oncogenic-driven mechanism of protein up-regulation.
Keywords: alternative TSS; cancer; transcription; translation; translation efficiency.
Conflict of interest statement
The authors thank Dr. Paola Signorelli (Department of Health Sciences, University of Milan, Milan, Italy) and Dr. Daniella Ishimaru (Medical University of South Carolina) for expert advice. The authors also thank Dr. Can Senkal and Dr. Yusuf Hannun (both of the Department of Medicine and Cancer Center at Stony Brook University) for critical input when writing the manuscript. The authors thank the Stony Brook DNA Sequencing facility for timely assistance. This work was supported by U.S. National Institutes of Health, National Cancer Institute Grant P01-CA097132 (to C. L. for Project #4) and the Stony Brook Scholars in Biomedical Sciences Award (to S. M.). The authors declare no conflicts of interest.
Figures




Similar articles
-
Regulation of human sphingomyelin synthase 1 translation through its 5'-untranslated region.FEBS Lett. 2020 Nov;594(22):3751-3764. doi: 10.1002/1873-3468.13952. Epub 2020 Oct 31. FEBS Lett. 2020. PMID: 33037626 Free PMC article.
-
Sphingomyelin synthase 1 activity is regulated by the BCR-ABL oncogene.J Lipid Res. 2013 Mar;54(3):794-805. doi: 10.1194/jlr.M033985. Epub 2012 Nov 16. J Lipid Res. 2013. PMID: 23160178 Free PMC article.
-
BCR-ABL/GATA1/miR-138 mini circuitry contributes to the leukemogenesis of chronic myeloid leukemia.Oncogene. 2014 Jan 2;33(1):44-54. doi: 10.1038/onc.2012.557. Epub 2012 Dec 3. Oncogene. 2014. PMID: 23208504
-
Human sphingomyelin synthase 1 gene (SMS1): organization, multiple mRNA splice variants and expression in adult tissues.Gene. 2011 Aug 1;481(2):65-75. doi: 10.1016/j.gene.2011.04.010. Epub 2011 Apr 29. Gene. 2011. PMID: 21549185
-
Transcription factor BACH2 is transcriptionally regulated by the BCR/ABL oncogene.Genes Chromosomes Cancer. 2001 Dec;32(4):353-63. doi: 10.1002/gcc.1200. Genes Chromosomes Cancer. 2001. PMID: 11746976
Cited by
-
Regulation of human sphingomyelin synthase 1 translation through its 5'-untranslated region.FEBS Lett. 2020 Nov;594(22):3751-3764. doi: 10.1002/1873-3468.13952. Epub 2020 Oct 31. FEBS Lett. 2020. PMID: 33037626 Free PMC article.
-
Cholesterol and Sphingolipid Enriched Lipid Rafts as Therapeutic Targets in Cancer.Int J Mol Sci. 2021 Jan 13;22(2):726. doi: 10.3390/ijms22020726. Int J Mol Sci. 2021. PMID: 33450869 Free PMC article. Review.
-
Ceramide Transfer Protein (CERT): An Overlooked Molecular Player in Cancer.Int J Mol Sci. 2021 Dec 7;22(24):13184. doi: 10.3390/ijms222413184. Int J Mol Sci. 2021. PMID: 34947980 Free PMC article. Review.
-
Ceramide/Sphingomyelin Rheostat Regulated by Sphingomyelin Synthases and Chronic Diseases in Murine Models.J Lipid Atheroscler. 2020 Sep;9(3):380-405. doi: 10.12997/jla.2020.9.3.380. Epub 2020 Jul 29. J Lipid Atheroscler. 2020. PMID: 33024732 Free PMC article. Review.
-
Sphingomyelin synthase 2 but not sphingomyelin synthase 1 is upregulated in ovarian cancer and involved in migration, growth and survival via different mechanisms.Am J Transl Res. 2021 May 15;13(5):4412-4421. eCollection 2021. Am J Transl Res. 2021. PMID: 34150023 Free PMC article.
References
-
- Yamaoka S., Miyaji M., Kitano T., Umehara H., Okazaki T. (2004) Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J. Biol. Chem. 279, 18688–18693 - PubMed
-
- Bernert J. T., Jr., Ullman M. D. (1981) Biosynthesis of sphingomyelin from erythro-ceramides and phosphatidylcholine by a microsomal cholinephosphotransferase. Biochim. Biophys. Acta 666, 99–109 - PubMed
-
- Hatch G. M., Vance D. E. (1992) Stimulation of sphingomyelin biosynthesis by brefeldin A and sphingomyelin breakdown by okadaic acid treatment of rat hepatocytes. J. Biol. Chem. 267, 12443–12451 - PubMed
-
- Marggraf W. D., Anderer F. A., Kanfer J. N. (1981) The formation of sphingomyelin from phosphatidylcholine in plasma membrane preparations from mouse fibroblasts. Biochim. Biophys. Acta 664, 61–73 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

