Loss of KDM6A Activates Super-Enhancers to Induce Gender-Specific Squamous-like Pancreatic Cancer and Confers Sensitivity to BET Inhibitors
- PMID: 29533787
- PMCID: PMC5854186
- DOI: 10.1016/j.ccell.2018.02.003
Loss of KDM6A Activates Super-Enhancers to Induce Gender-Specific Squamous-like Pancreatic Cancer and Confers Sensitivity to BET Inhibitors
Abstract
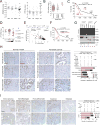
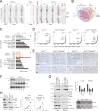
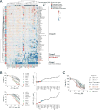
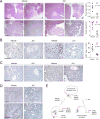
KDM6A, an X chromosome-encoded histone demethylase and member of the COMPASS-like complex, is frequently mutated in a broad spectrum of malignancies and contributes to oncogenesis with poorly characterized mechanisms. We found that KDM6A loss induced squamous-like, metastatic pancreatic cancer selectively in females through deregulation of the COMPASS-like complex and aberrant activation of super-enhancers regulating ΔNp63, MYC, and RUNX3 oncogenes. This subtype of tumor developed in males had concomitant loss of UTY and KDM6A, suggesting overlapping roles, and points to largely demethylase independent tumor suppressor functions. We also demonstrate that KDM6A-deficient pancreatic cancer is selectively sensitive to BET inhibitors, which reversed squamous differentiation and restrained tumor growth in vivo, highlighting a therapeutic niche for patient tailored therapies.
Keywords: COMPASS-like complex; JQ1; KDM6A; KMT2D; MYC; UTY; p63; pancreatic cancer; squamous; super-enhancer.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Pancreatic cancer: The COMPASS shows the way.Nat Rev Cancer. 2018 May;18(5):373. doi: 10.1038/nrc.2018.29. Epub 2018 Apr 3. Nat Rev Cancer. 2018. PMID: 29610488 No abstract available.
-
Pancreatic cancer foiled by a switch of tumour subtype.Nature. 2018 May;557(7706):500-501. doi: 10.1038/d41586-018-05129-6. Nature. 2018. PMID: 29777189 No abstract available.
References
-
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. - PubMed
-
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

