Circadian clock component REV-ERBα controls homeostatic regulation of pulmonary inflammation
- PMID: 29533925
- PMCID: PMC5983347
- DOI: 10.1172/JCI93910
Circadian clock component REV-ERBα controls homeostatic regulation of pulmonary inflammation
Abstract
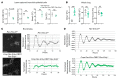
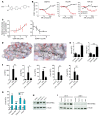
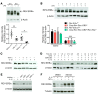
Recent studies reveal that airway epithelial cells are critical pulmonary circadian pacemaker cells, mediating rhythmic inflammatory responses. Using mouse models, we now identify the rhythmic circadian repressor REV-ERBα as essential to the mechanism coupling the pulmonary clock to innate immunity, involving both myeloid and bronchial epithelial cells in temporal gating and determining amplitude of response to inhaled endotoxin. Dual mutation of REV-ERBα and its paralog REV-ERBβ in bronchial epithelia further augmented inflammatory responses and chemokine activation, but also initiated a basal inflammatory state, revealing a critical homeostatic role for REV-ERB proteins in the suppression of the endogenous proinflammatory mechanism in unchallenged cells. However, REV-ERBα plays the dominant role, as deletion of REV-ERBβ alone had no impact on inflammatory responses. In turn, inflammatory challenges cause striking changes in stability and degradation of REV-ERBα protein, driven by SUMOylation and ubiquitination. We developed a novel selective oxazole-based inverse agonist of REV-ERB, which protects REV-ERBα protein from degradation, and used this to reveal how proinflammatory cytokines trigger rapid degradation of REV-ERBα in the elaboration of an inflammatory response. Thus, dynamic changes in stability of REV-ERBα protein couple the core clock to innate immunity.
Keywords: Inflammation; Innate immunity; Mouse models; Neutrophils; Pulmonology.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

