Insights into the Evolution of Host Association through the Isolation and Characterization of a Novel Human Periodontal Pathobiont, Desulfobulbus oralis
- PMID: 29535201
- PMCID: PMC5850319
- DOI: 10.1128/mBio.02061-17
Insights into the Evolution of Host Association through the Isolation and Characterization of a Novel Human Periodontal Pathobiont, Desulfobulbus oralis
Abstract
The human oral microbiota encompasses representatives of many bacterial lineages that have not yet been cultured. Here we describe the isolation and characterization of previously uncultured Desulfobulbus oralis, the first human-associated representative of its genus. As mammalian-associated microbes rarely have free-living close relatives, D. oralis provides opportunities to study how bacteria adapt and evolve within a host. This sulfate-reducing deltaproteobacterium has adapted to the human oral subgingival niche by curtailing its physiological repertoire, losing some biosynthetic abilities and metabolic independence, and by dramatically reducing environmental sensing and signaling capabilities. The genes that enable free-living Desulfobulbus to synthesize the potent neurotoxin methylmercury were also lost by D. oralis, a notably positive outcome of host association. However, horizontal gene acquisitions from other members of the microbiota provided novel mechanisms of interaction with the human host, including toxins like leukotoxin and hemolysins. Proteomic and transcriptomic analysis revealed that most of those factors are actively expressed, including in the subgingival environment, and some are secreted. Similar to other known oral pathobionts, D. oralis can trigger a proinflammatory response in oral epithelial cells, suggesting a direct role in the development of periodontal disease.IMPORTANCE Animal-associated microbiota likely assembled as a result of numerous independent colonization events by free-living microbes followed by coevolution with their host and other microbes. Through specific adaptation to various body sites and physiological niches, microbes have a wide range of contributions, from beneficial to disease causing. Desulfobulbus oralis provides insights into genomic and physiological transformations associated with transition from an open environment to a host-dependent lifestyle and the emergence of pathogenicity. Through a multifaceted mechanism triggering a proinflammatory response, D. oralis is a novel periodontal pathobiont. Even though culture-independent approaches can provide insights into the potential role of the human microbiome "dark matter," cultivation and experimental characterization remain important to studying the roles of individual organisms in health and disease.
Keywords: evolution; genome analysis; oral microbiology; periodontitis; proteomics.
Figures
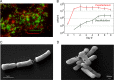
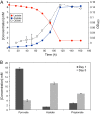
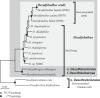


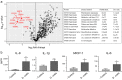
References
-
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10:497–506. doi: 10.1016/j.chom.2011.10.006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
