Identification and characterisation of a cryptic Golgi complex in Naegleria gruberi
- PMID: 29535209
- PMCID: PMC5963838
- DOI: 10.1242/jcs.213306
Identification and characterisation of a cryptic Golgi complex in Naegleria gruberi
Abstract
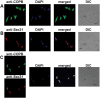
Although the Golgi complex has a conserved morphology of flattened stacked cisternae in most eukaryotes, it has lost the stacked organisation in several lineages, raising the question of what range of morphologies is possible for the Golgi. In order to understand this diversity, it is necessary to characterise the Golgi in many different lineages. Here, we identify the Golgi complex in Naegleria, one of the first descriptions of an unstacked Golgi organelle in a non-parasitic eukaryote, other than fungi. We provide a comprehensive list of Golgi-associated membrane trafficking genes encoded in two species of Naegleria and show that nearly all are expressed in mouse-passaged N. fowleri cells. We then study distribution of the Golgi marker (Ng)CopB by fluorescence in Naegleria gruberi, identifying membranous structures that are disrupted by Brefeldin A treatment, consistent with Golgi localisation. Confocal and immunoelectron microscopy reveals that NgCOPB localises to tubular membranous structures. Our data identify the Golgi organelle for the first time in this major eukaryotic lineage, and provide the rare example of a tubular morphology, representing an important sampling point for the comparative understanding of Golgi organellar diversity.This article has an associated First Person interview with the first author of the paper.
Keywords: Brefeldin A; COPI; Dictyosome; Evolutionary cell biology; Membrane trafficking; Protist.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures



Similar articles
-
A sophisticated, differentiated Golgi in the ancestor of eukaryotes.BMC Biol. 2018 Mar 7;16(1):27. doi: 10.1186/s12915-018-0492-9. BMC Biol. 2018. PMID: 29510703 Free PMC article.
-
The genome of Naegleria gruberi illuminates early eukaryotic versatility.Cell. 2010 Mar 5;140(5):631-42. doi: 10.1016/j.cell.2010.01.032. Cell. 2010. PMID: 20211133
-
Live-cell imaging of dual-labeled Golgi stacks in tobacco BY-2 cells reveals similar behaviors for different cisternae during movement and brefeldin A treatment.Mol Plant. 2011 Sep;4(5):896-908. doi: 10.1093/mp/ssr067. Epub 2011 Aug 25. Mol Plant. 2011. PMID: 21873295
-
Evolution and diversity of the Golgi body.FEBS Lett. 2009 Dec 3;583(23):3738-45. doi: 10.1016/j.febslet.2009.10.025. Epub 2009 Oct 20. FEBS Lett. 2009. PMID: 19837068 Review.
-
The Naegleria genome: a free-living microbial eukaryote lends unique insights into core eukaryotic cell biology.Res Microbiol. 2011 Jul-Aug;162(6):607-18. doi: 10.1016/j.resmic.2011.03.003. Epub 2011 Mar 21. Res Microbiol. 2011. PMID: 21392573 Free PMC article. Review.
Cited by
-
Noninvasive diagnostic biomarkers, genomic profiling, and advanced microscopic imaging in the early detection and characterization of Naegleria fowleri infections leading to primary amebic meningoencephalitis (PAM).Ann Med Surg (Lond). 2024 Feb 28;86(4):2032-2048. doi: 10.1097/MS9.0000000000001843. eCollection 2024 Apr. Ann Med Surg (Lond). 2024. PMID: 38576920 Free PMC article. Review.
-
Localization and functional characterization of the alternative oxidase in Naegleria.J Eukaryot Microbiol. 2022 Jul;69(4):e12908. doi: 10.1111/jeu.12908. Epub 2022 May 3. J Eukaryot Microbiol. 2022. PMID: 35322502 Free PMC article.
References
-
- Beznoussenko G. V., Dolgikh V. V., Seliverstova E. V., Semenov P. B., Tokarev Y. S., Trucco A., Micaroni M., Di Giandomenico D., Auinger P., Senderskiy I. V. et al. (2007). Analogs of the Golgi complex in microsporidia: structure and avesicular mechanisms of function. 120, 1288-1298. 10.1242/jcs.03402 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

