Pre-equilibrium competitive library screening for tuning inhibitor association rate and specificity toward serine proteases
- PMID: 29535275
- PMCID: PMC5929103
- DOI: 10.1042/BCJ20180070
Pre-equilibrium competitive library screening for tuning inhibitor association rate and specificity toward serine proteases
Abstract
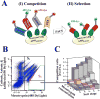
High structural and sequence similarity within protein families can pose significant challenges to the development of selective inhibitors, especially toward proteolytic enzymes. Such enzymes usually belong to large families of closely similar proteases and may also hydrolyze, with different rates, protein- or peptide-based inhibitors. To address this challenge, we employed a combinatorial yeast surface display library approach complemented with a novel pre-equilibrium, competitive screening strategy for facile assessment of the effects of multiple mutations on inhibitor association rates and binding specificity. As a proof of principle for this combined approach, we utilized this strategy to alter inhibitor/protease association rates and to tailor the selectivity of the amyloid β-protein precursor Kunitz protease inhibitor domain (APPI) for inhibition of the oncogenic protease mesotrypsin, in the presence of three competing serine proteases, anionic trypsin, cationic trypsin and kallikrein-6. We generated a variant, designated APPIP13W/M17G/I18F/F34V, with up to 30-fold greater specificity relative to the parental APPIM17G/I18F/F34V protein, and 6500- to 230 000-fold improved specificity relative to the wild-type APPI protein in the presence of the other proteases tested. A series of molecular docking simulations suggested a mechanism of interaction that supported the biochemical results. These simulations predicted that the selectivity and specificity are affected by the interaction of the mutated APPI residues with nonconserved enzyme residues located in or near the binding site. Our strategy will facilitate a better understanding of the binding landscape of multispecific proteins and will pave the way for design of new drugs and diagnostic tools targeting proteases and other proteins.
Keywords: directed evolution; protease inhibitor; protein engineering; protein–protein interactions (PPIs); serine proteases.
© 2018 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
The authors declare that they have no conflict of interest with respect to publication of this paper.
Figures



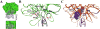
Similar articles
-
Combinatorial protein engineering of proteolytically resistant mesotrypsin inhibitors as candidates for cancer therapy.Biochem J. 2016 May 15;473(10):1329-41. doi: 10.1042/BJ20151410. Epub 2016 Mar 8. Biochem J. 2016. PMID: 26957636 Free PMC article.
-
Sequence and conformational specificity in substrate recognition: several human Kunitz protease inhibitor domains are specific substrates of mesotrypsin.J Biol Chem. 2014 Nov 21;289(47):32783-97. doi: 10.1074/jbc.M114.609560. Epub 2014 Oct 9. J Biol Chem. 2014. PMID: 25301953 Free PMC article.
-
The amyloid precursor protein/protease nexin 2 Kunitz inhibitor domain is a highly specific substrate of mesotrypsin.J Biol Chem. 2010 Jan 15;285(3):1939-49. doi: 10.1074/jbc.M109.057216. Epub 2009 Nov 17. J Biol Chem. 2010. PMID: 19920152 Free PMC article.
-
Mechanism-based isocoumarin inhibitors for serine proteases: use of active site structure and substrate specificity in inhibitor design.J Cell Biochem. 1989 Jan;39(1):33-46. doi: 10.1002/jcb.240390105. J Cell Biochem. 1989. PMID: 2654146 Review.
-
Conformational homogeneity in molecular recognition by proteolytic enzymes.J Mol Recognit. 1999 Nov-Dec;12(6):363-70. doi: 10.1002/(SICI)1099-1352(199911/12)12:6<363::AID-JMR478>3.0.CO;2-M. J Mol Recognit. 1999. PMID: 10611646 Review.
Cited by
-
Quantitative mapping of binding specificity landscapes for homologous targets by using a high-throughput method.Biochem J. 2020 May 15;477(9):1701-1719. doi: 10.1042/BCJ20200188. Biochem J. 2020. PMID: 32296833 Free PMC article.
-
Climbing Up and Down Binding Landscapes through Deep Mutational Scanning of Three Homologous Protein-Protein Complexes.J Am Chem Soc. 2021 Oct 20;143(41):17261-17275. doi: 10.1021/jacs.1c08707. Epub 2021 Oct 5. J Am Chem Soc. 2021. PMID: 34609866 Free PMC article.
-
Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.J Biol Chem. 2019 Mar 29;294(13):5105-5120. doi: 10.1074/jbc.RA118.007292. Epub 2019 Jan 30. J Biol Chem. 2019. PMID: 30700553 Free PMC article.
-
Discovery of an autoinhibited conformation in mesotrypsin reveals a strategy for selective serine protease inhibition.Sci Adv. 2025 Jul 4;11(27):eadu9129. doi: 10.1126/sciadv.adu9129. Epub 2025 Jul 4. Sci Adv. 2025. PMID: 40614191 Free PMC article.
-
PRSS3/Mesotrypsin and kallikrein-related peptidase 5 are associated with poor prognosis and contribute to tumor cell invasion and growth in lung adenocarcinoma.Sci Rep. 2019 Feb 12;9(1):1844. doi: 10.1038/s41598-018-38362-0. Sci Rep. 2019. PMID: 30755669 Free PMC article.
References
-
- Koblinski JE, Ahram M, Sloane BF. Unraveling the role of proteases in cancer. Clinica Chimica Acta. 2000;291:113–135. - PubMed
-
- Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nature reviews Cancer. 2002;2:161–174. - PubMed
-
- Radisky ES, Radisky DC. Stromal induction of breast cancer: inflammation and invasion. Reviews in endocrine & metabolic disorders. 2007;8:279–287. - PubMed
-
- Turk B. Targeting proteases: successes, failures and future prospects. Nature Reviews: Drug Discovery. 2006;5:785–799. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

