Serine Hydroxymethyltransferase ShrA (PA2444) Controls Rugose Small-Colony Variant Formation in Pseudomonas aeruginosa
- PMID: 29535691
- PMCID: PMC5835335
- DOI: 10.3389/fmicb.2018.00315
Serine Hydroxymethyltransferase ShrA (PA2444) Controls Rugose Small-Colony Variant Formation in Pseudomonas aeruginosa
Abstract
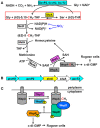
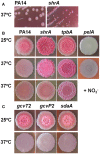
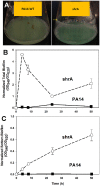
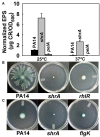
Pseudomonas aeruginosa causes many biofilm infections, and the rugose small-colony variants (RSCVs) of this bacterium are important for infection. We found here that inactivation of PA2444, which we determined to be a serine hydroxymethyltransferase (SHMT), leads to the RSCV phenotype of P. aeruginosa PA14. In addition, loss of PA2444 increases biofilm formation by two orders of magnitude, increases exopolysaccharide by 45-fold, and abolishes swarming. The RSCV phenotype is related to higher cyclic diguanylate concentrations due to increased activity of the Wsp chemosensory system, including diguanylate cyclase WspR. By characterizing the PA2444 enzyme in vitro, we determined the physiological function of PA2444 protein by relating it to S-adenosylmethionine (SAM) concentrations and methylation of a membrane bound methyl-accepting chemotaxis protein WspA. A whole transcriptome analysis also revealed PA2444 is related to the redox state of the cells, and the altered redox state was demonstrated by an increase in the intracellular NADH/NAD+ ratio. Hence, we provide a mechanism for how an enzyme of central metabolism controls the community behavior of the bacterium, and suggest the PA2444 protein should be named ShrA for serine hydroxymethyltransferase related to rugose colony formation.
Keywords: Pseudomonas aeruginosa; biofilm formation; rugose; serine hydroxymethyltransferase; small colony variants.
Figures



Similar articles
-
Pseudomonas aeruginosa Interstrain Dynamics and Selection of Hyperbiofilm Mutants during a Chronic Infection.mBio. 2019 Aug 13;10(4):e01698-19. doi: 10.1128/mBio.01698-19. mBio. 2019. PMID: 31409682 Free PMC article.
-
Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung.J Bacteriol. 2009 Jun;191(11):3492-503. doi: 10.1128/JB.00119-09. Epub 2009 Mar 27. J Bacteriol. 2009. PMID: 19329647 Free PMC article.
-
Mutations in surface-sensing receptor WspA lock the Wsp signal transduction system into a constitutively active state.Environ Microbiol. 2022 Mar;24(3):1150-1165. doi: 10.1111/1462-2920.15763. Epub 2021 Sep 20. Environ Microbiol. 2022. PMID: 34499799
-
Rugose small colony variant and its hyper-biofilm in Pseudomonas aeruginosa: Adaption, evolution, and biotechnological potential.Biotechnol Adv. 2021 Dec;53:107862. doi: 10.1016/j.biotechadv.2021.107862. Epub 2021 Oct 28. Biotechnol Adv. 2021. PMID: 34718136 Review.
-
Small colony variants of Pseudomonas aeruginosa in chronic bacterial infection of the lung in cystic fibrosis.Future Microbiol. 2015;10(2):231-9. doi: 10.2217/fmb.14.107. Future Microbiol. 2015. PMID: 25689535 Review.
Cited by
-
The sulfur-related metabolic status of Aspergillus fumigatus during infection reveals cytosolic serine hydroxymethyltransferase as a promising antifungal target.Virulence. 2025 Dec;16(1):2449075. doi: 10.1080/21505594.2024.2449075. Epub 2025 Jan 17. Virulence. 2025. PMID: 39825596 Free PMC article.
-
Comparative Proteomic Analysis of Protein Patterns of Stenotrophomonas maltophilia in Biofilm and Planktonic Lifestyles.Microorganisms. 2023 Feb 9;11(2):442. doi: 10.3390/microorganisms11020442. Microorganisms. 2023. PMID: 36838406 Free PMC article.
-
Identifying potential inhibitors of biofilm-antagonistic proteins to promote biofilm formation: a virtual screening and molecular dynamics simulations approach.Mol Divers. 2022 Aug;26(4):2135-2147. doi: 10.1007/s11030-021-10320-5. Epub 2021 Sep 21. Mol Divers. 2022. PMID: 34546549
-
Pseudomonas aeruginosa reference strains PAO1 and PA14: A genomic, phenotypic, and therapeutic review.Front Microbiol. 2022 Oct 13;13:1023523. doi: 10.3389/fmicb.2022.1023523. eCollection 2022. Front Microbiol. 2022. PMID: 36312971 Free PMC article. Review.
-
A specific innate immune response silences the virulence of Pseudomonas aeruginosa in a latent infection model in the Drosophila melanogaster host.PLoS Pathog. 2024 Jun 4;20(6):e1012252. doi: 10.1371/journal.ppat.1012252. eCollection 2024 Jun. PLoS Pathog. 2024. PMID: 38833496 Free PMC article.
References
-
- Berg J. M., Tymoczko J. L., Stryer L. (2002). Biochemistry. New York, NY: W. H. Freeman and Company.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

