Epigenomic Modifications Mediating Antibody Maturation
- PMID: 29535729
- PMCID: PMC5834911
- DOI: 10.3389/fimmu.2018.00355
Epigenomic Modifications Mediating Antibody Maturation
Abstract
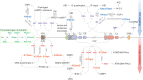
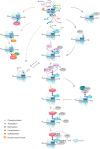
Epigenetic modifications, such as histone modifications, DNA methylation status, and non-coding RNAs (ncRNA), all contribute to antibody maturation during somatic hypermutation (SHM) and class-switch recombination (CSR). Histone modifications alter the chromatin landscape and, together with DNA primary and tertiary structures, they help recruit Activation-Induced Cytidine Deaminase (AID) to the immunoglobulin (Ig) locus. AID is a potent DNA mutator, which catalyzes cytosine-to-uracil deamination on single-stranded DNA to create U:G mismatches. It has been shown that alternate chromatin modifications, in concert with ncRNAs and potentially DNA methylation, regulate AID recruitment and stabilize DNA repair factors. We, hereby, assess the combination of these distinct modifications and discuss how they contribute to initiating differential DNA repair pathways at the Ig locus, which ultimately leads to enhanced antibody-antigen binding affinity (SHM) or antibody isotype switching (CSR). We will also highlight how misregulation of epigenomic regulation during DNA repair can compromise antibody development and lead to a number of immunological syndromes and cancer.
Keywords: B cell maturation; antibody diversity; class-switch recombination; cytosine deamination; epigenetic modifications; epigenomics and epigenetics; somatic hypermutation.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

