NLRX1 Modulates Immunometabolic Mechanisms Controlling the Host-Gut Microbiota Interactions during Inflammatory Bowel Disease
- PMID: 29535731
- PMCID: PMC5834749
- DOI: 10.3389/fimmu.2018.00363
NLRX1 Modulates Immunometabolic Mechanisms Controlling the Host-Gut Microbiota Interactions during Inflammatory Bowel Disease
Abstract
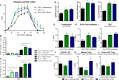
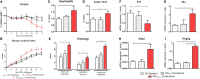
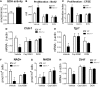
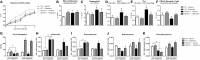
Interactions among the gut microbiome, dysregulated immune responses, and genetic factors contribute to the pathogenesis of inflammatory bowel disease (IBD). Nlrx1-/- mice have exacerbated disease severity, colonic lesions, and increased inflammatory markers. Global transcriptomic analyses demonstrate enhanced mucosal antimicrobial defense response, chemokine and cytokine expression, and epithelial cell metabolism in colitic Nlrx1-/- mice compared to wild-type (WT) mice. Cell-specificity studies using cre-lox mice demonstrate that the loss of NLRX1 in intestinal epithelial cells (IEC) recapitulate the increased sensitivity to DSS colitis observed in whole body Nlrx1-/- mice. Further, organoid cultures of Nlrx1-/- and WT epithelial cells confirm the altered patterns of proliferation, amino acid metabolism, and tight junction expression. These differences in IEC behavior can impact the composition of the microbiome. Microbiome analyses demonstrate that colitogenic bacterial taxa such as Veillonella and Clostridiales are increased in abundance in Nlrx1-/- mice and in WT mice co-housed with Nlrx1-/- mice. The transfer of an Nlrx1-/--associated gut microbiome through co-housing worsens disease in WT mice confirming the contributions of the microbiome to the Nlrx1-/- phenotype. To validate NLRX1 effects on IEC metabolism mediate gut-microbiome interactions, restoration of WT glutamine metabolic profiles through either exogenous glutamine supplementation or administration of 6-diazo-5-oxo-l-norleucine abrogates differences in inflammation, microbiome, and overall disease severity in Nlrx1-/- mice. The influence NLRX1 deficiency on SIRT1-mediated effects is identified to be an upstream controller of the Nlrx1-/- phenotype in intestinal epithelial cell function and metabolism. The altered IEC function and metabolisms leads to changes in barrier permeability and microbiome interactions, in turn, promoting greater translocation and inflammation and resulting in an increased disease severity. In conclusion, NLRX1 is an immunoregulatory molecule and a candidate modulator of the interplay between mucosal inflammation, metabolism, and the gut microbiome during IBD.
Keywords: NLRX1; gut microbiome; immunometabolism; inflammatory bowel disease; intestinal epithelial cells; mucosal immunology.
Figures



Similar articles
-
NLRX1 Regulates Effector and Metabolic Functions of CD4+ T Cells.J Immunol. 2017 Mar 15;198(6):2260-2268. doi: 10.4049/jimmunol.1601547. Epub 2017 Feb 3. J Immunol. 2017. PMID: 28159898 Free PMC article.
-
Fermented barley and soybean (BS) mixture enhances intestinal barrier function in dextran sulfate sodium (DSS)-induced colitis mouse model.BMC Complement Altern Med. 2016 Dec 3;16(1):498. doi: 10.1186/s12906-016-1479-0. BMC Complement Altern Med. 2016. PMID: 27912750 Free PMC article.
-
Enteric Delivery of Regenerating Family Member 3 alpha Alters the Intestinal Microbiota and Controls Inflammation in Mice With Colitis.Gastroenterology. 2018 Mar;154(4):1009-1023.e14. doi: 10.1053/j.gastro.2017.11.003. Epub 2017 Nov 11. Gastroenterology. 2018. PMID: 29133078
-
Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis.J Biomed Sci. 2018 Nov 9;25(1):79. doi: 10.1186/s12929-018-0483-8. J Biomed Sci. 2018. PMID: 30413188 Free PMC article. Review.
-
Roles of intestinal epithelial cells in the maintenance of gut homeostasis.Exp Mol Med. 2017 May 26;49(5):e338. doi: 10.1038/emm.2017.20. Exp Mol Med. 2017. PMID: 28546564 Free PMC article. Review.
Cited by
-
Insights into the structure of NLR family member X1: Paving the way for innovative drug discovery.Comput Struct Biotechnol J. 2024 Sep 22;23:3506-3513. doi: 10.1016/j.csbj.2024.09.013. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39435340 Free PMC article.
-
Co-Housing and Fecal Microbiota Transplantation: Technical Support for TCM Herbal Treatment of Extra-Intestinal Diseases Based on Gut Microbial Ecosystem Remodeling.Drug Des Devel Ther. 2023 Dec 24;17:3803-3831. doi: 10.2147/DDDT.S443462. eCollection 2023. Drug Des Devel Ther. 2023. PMID: 38155743 Free PMC article. Review.
-
NAD+ Metabolism and Immune Regulation: New Approaches to Inflammatory Bowel Disease Therapies.Antioxidants (Basel). 2023 Jun 7;12(6):1230. doi: 10.3390/antiox12061230. Antioxidants (Basel). 2023. PMID: 37371959 Free PMC article. Review.
-
Focusing on the Cell Type Specific Regulatory Actions of NLRX1.Int J Mol Sci. 2021 Jan 28;22(3):1316. doi: 10.3390/ijms22031316. Int J Mol Sci. 2021. PMID: 33525671 Free PMC article. Review.
-
NLRX1 limits inflammatory neurodegeneration in the anterior visual pathway.J Neuroinflammation. 2025 Jan 28;22(1):21. doi: 10.1186/s12974-025-03339-0. J Neuroinflammation. 2025. PMID: 39875919 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

