Epstein-Barr virus stably confers an invasive phenotype to epithelial cells through reprogramming of the WNT pathway
- PMID: 29535816
- PMCID: PMC5828208
- DOI: 10.18632/oncotarget.23824
Epstein-Barr virus stably confers an invasive phenotype to epithelial cells through reprogramming of the WNT pathway
Abstract
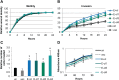
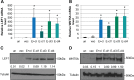
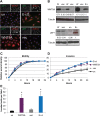
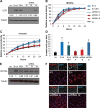
Epstein-Barr virus (EBV)-associated carcinomas, such as nasopharyngeal carcinoma (NPC), exhibit an undifferentiated and metastatic phenotype. To determine viral contributions involved in the invasive phenotype of EBV-associated carcinomas, EBV-infected human telomerase-immortalized normal oral keratinocytes (NOK) were investigated. EBV-infected NOK were previously shown to undergo epigenetic reprogramming involving CpG island hypermethylation and delayed responsiveness to differentiation. Here, we show that EBV-infected NOK acquired an invasive phenotype that was epigenetically retained after viral loss. The transcription factor lymphoid enhancer factor 1 (LEF1) and the secreted ligand WNT5A, expressed in NPC, were increased in EBV-infected NOK with sustained expression for more than 20 passages after viral loss. Increased LEF1 levels involved four LEF1 variants, and EBV-infected NOK showed a lack of responsiveness to β-catenin activation. Although forced expression of WNT5A and LEF1 enhanced the invasiveness of parental NOK, LEF1 knockdown reversed the invasive phenotype of EBV-infected NOK in the presence of WNT5A. Viral reprogramming of LEF1 and WNT5A was observed several passages after EBV infection, suggesting that LEF1 and WNT5A may provide a selective advantage to virally-infected cells. Our findings suggest that EBV epigenetically reprogrammed epithelial cells with features of basal, wound healing keratinocytes, with LEF1 contributing to the metastatic phenotype of EBV-associated carcinomas.
Keywords: Epstein-Barr virus; LEF1; WNT; epigenetics; invasion.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no conflicts of interests
Figures


Similar articles
-
Genome-wide DNA methylation as an epigenetic consequence of Epstein-Barr virus infection of immortalized keratinocytes.J Virol. 2014 Oct;88(19):11442-58. doi: 10.1128/JVI.00972-14. Epub 2014 Jul 23. J Virol. 2014. PMID: 25056883 Free PMC article.
-
A distinct isoform of lymphoid enhancer binding factor 1 (LEF1) epigenetically restricts EBV reactivation to maintain viral latency.PLoS Pathog. 2023 Dec 19;19(12):e1011873. doi: 10.1371/journal.ppat.1011873. eCollection 2023 Dec. PLoS Pathog. 2023. PMID: 38113273 Free PMC article.
-
Epstein-Barr virus infection in immortalized nasopharyngeal epithelial cells: regulation of infection and phenotypic characterization.Int J Cancer. 2010 Oct 1;127(7):1570-83. doi: 10.1002/ijc.25173. Int J Cancer. 2010. PMID: 20091869
-
Epstein-Barr virus infection and the pathogenesis of nasopharyngeal carcinoma: viral gene expression, tumour cell phenotype, and the role of the lymphoid stroma.Semin Cancer Biol. 1996 Aug;7(4):165-74. doi: 10.1006/scbi.1996.0023. Semin Cancer Biol. 1996. PMID: 8946600 Review.
-
Epigenetic Alterations in Epstein-Barr Virus-Associated Diseases.Adv Exp Med Biol. 2016;879:39-69. doi: 10.1007/978-3-319-24738-0_3. Adv Exp Med Biol. 2016. PMID: 26659263 Review.
Cited by
-
Viruses in colorectal cancer.Mol Oncol. 2022 Apr;16(7):1423-1450. doi: 10.1002/1878-0261.13100. Epub 2021 Sep 30. Mol Oncol. 2022. PMID: 34514694 Free PMC article. Review.
-
Epstein-Barr Virus Infection in Lung Cancer: Insights and Perspectives.Pathogens. 2022 Jan 21;11(2):132. doi: 10.3390/pathogens11020132. Pathogens. 2022. PMID: 35215076 Free PMC article. Review.
-
Type 1 and Type 2 Epstein-Barr viruses induce proliferation, and inhibit differentiation, in infected telomerase-immortalized normal oral keratinocytes.PLoS Pathog. 2022 Oct 3;18(10):e1010868. doi: 10.1371/journal.ppat.1010868. eCollection 2022 Oct. PLoS Pathog. 2022. PMID: 36190982 Free PMC article.
-
Transcriptome reprogramming of Epstein-Barr virus infected epithelial and B cells reveals distinct host-virus interaction profiles.Cell Death Dis. 2022 Oct 22;13(10):894. doi: 10.1038/s41419-022-05327-1. Cell Death Dis. 2022. PMID: 36272970 Free PMC article.
-
Epstein-Barr Virus Infection Promotes Epithelial Cell Growth by Attenuating Differentiation-Dependent Exit from the Cell Cycle.mBio. 2019 Aug 20;10(4):e01332-19. doi: 10.1128/mBio.01332-19. mBio. 2019. PMID: 31431547 Free PMC article.
References
-
- Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365:2041–54. https://doi.org/10.1016/S0140-6736(05)66698-6 - DOI - PubMed
-
- Chan AS, To KF, Lo KW, Mak KF, Pak W, Chiu B, Tse GM, Ding M, Li X, Lee JC, Huang DP. High frequency of chromosome 3p deletion in histologically normal nasopharyngeal epithelia from southern Chinese. Cancer Res. 2000;60:5365–70. - PubMed
-
- Henle W, Henle G. Evidence for a relation of Epstein-Barr virus to Burkitt's lymphoma and nasopharyngeal carcinoma. Bibl Haematol. 1970. pp. 706–13. - PubMed
-
- Pathmanathan R, Prasad U, Sadler R, Flynn K, Raab-Traub N. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N Engl J Med. 1995;333:693–8. https://doi.org/10.1056/NEJM199509143331103 - DOI - PubMed
-
- zur Hausen H, Schulte-Holthausen H, Klein G, Henle W, Henle G, Clifford P, Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056–8. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

