Detection of PRMT1 inhibitors with stopped flow fluorescence
- PMID: 29535867
- PMCID: PMC5843908
- DOI: 10.1038/s41392-018-0009-6
Detection of PRMT1 inhibitors with stopped flow fluorescence
Abstract
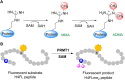
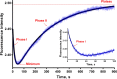
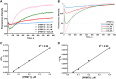
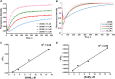
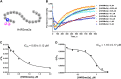
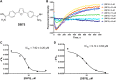
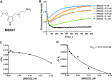
Protein arginine methyltransferases (PRMTs) are crucial epigenetic regulators in eukaryotic organisms that serve as histone writers for chromatin remodeling. PRMTs also methylate a variety of non-histone protein substrates to modulate their function and activity. The development of potent PRMT inhibitors has become an emerging and imperative research area in the drug discovery field to provide novel therapeutic agents for treating diseases and as tools to investigate the biological functions of PRMTs. PRMT1 is the major type I enzyme that catalyzes the formation of asymmetric dimethyl arginine, and PRMT1 plays important regulatory roles in signal transduction, transcriptional activation, RNA splicing, and DNA repair. Aberrant expression of PRMT1 is found in many types of cancers, pulmonary diseases, cardiovascular disease, diabetes, and renal diseases. PRMT1 is a highly promising target for therapeutic development. We created a stopped flow fluorescence-based assay for PRMT1 inhibitor detection and characterization that has the advantages of being homogeneous, nonradioactive, and mix-and-measure in nature, allowing for continuous measurement of the methylation reaction and its inhibition. To our knowledge, this is the first continuous assay for PRMT1 reaction detection and inhibitor characterization. The approach is not only capable of quantitatively determining the potency (IC50) of PRMT1 inhibitors but can also distinguish cofactor-competitive inhibitors, substrate-competitive inhibitors, and mixed-type inhibitors.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

