Insights into the Active Site of Coproheme Decarboxylase from Listeria monocytogenes
- PMID: 29536725
- PMCID: PMC5940323
- DOI: 10.1021/acs.biochem.8b00186
Insights into the Active Site of Coproheme Decarboxylase from Listeria monocytogenes
Abstract
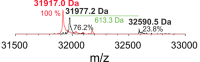
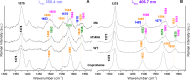
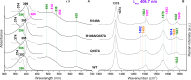
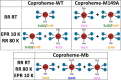
Coproheme decarboxylases (ChdC) catalyze the hydrogen peroxide-mediated conversion of coproheme to heme b. This work compares the structure and function of wild-type (WT) coproheme decarboxylase from Listeria monocytogenes and its M149A, Q187A, and M149A/Q187A mutants. The UV-vis, resonance Raman, and electron paramagnetic resonance spectroscopies clearly show that the ferric form of the WT protein is a pentacoordinate quantum mechanically mixed-spin state, which is very unusual in biological systems. Exchange of the Met149 residue to Ala dramatically alters the heme coordination, which becomes a 6-coordinate low spin species with the amide nitrogen atom of the Q187 residue bound to the heme iron. The interaction between M149 and propionyl 2 is found to play an important role in keeping the Q187 residue correctly positioned for closure of the distal cavity. This is confirmed by the observation that in the M149A variant two CO conformers are present corresponding to open (A0) and closed (A1) conformations. The CO of the latter species, the only conformer observed in the WT protein, is H-bonded to Q187. In the absence of the Q187 residue or in the adducts of all the heme b forms of ChdC investigated herein (containing vinyls in positions 2 and 4), only the A0 conformer has been found. Moreover, M149 is shown to be involved in the formation of a covalent bond with a vinyl substituent of heme b at excess of hydrogen peroxide.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
The hydrogen bonding network of coproheme in coproheme decarboxylase from Listeria monocytogenes: Effect on structure and catalysis.J Inorg Biochem. 2019 Jun;195:61-70. doi: 10.1016/j.jinorgbio.2019.03.009. Epub 2019 Mar 21. J Inorg Biochem. 2019. PMID: 30925402 Free PMC article.
-
Understanding molecular enzymology of porphyrin-binding α + β barrel proteins - One fold, multiple functions.Biochim Biophys Acta Proteins Proteom. 2021 Jan;1869(1):140536. doi: 10.1016/j.bbapap.2020.140536. Epub 2020 Sep 4. Biochim Biophys Acta Proteins Proteom. 2021. PMID: 32891739 Free PMC article. Review.
-
An active site at work - the role of key residues in C. diphteriae coproheme decarboxylase.J Inorg Biochem. 2022 Apr;229:111718. doi: 10.1016/j.jinorgbio.2022.111718. Epub 2022 Jan 6. J Inorg Biochem. 2022. PMID: 35051755
-
Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.FEBS J. 2016 Dec;283(23):4386-4401. doi: 10.1111/febs.13930. Epub 2016 Nov 14. FEBS J. 2016. PMID: 27758026 Free PMC article.
-
HemQ: An iron-coproporphyrin oxidative decarboxylase for protoheme synthesis in Firmicutes and Actinobacteria.Arch Biochem Biophys. 2015 May 15;574:27-35. doi: 10.1016/j.abb.2015.02.017. Epub 2015 Feb 21. Arch Biochem Biophys. 2015. PMID: 25711532 Free PMC article. Review.
Cited by
-
The hydrogen bonding network of coproheme in coproheme decarboxylase from Listeria monocytogenes: Effect on structure and catalysis.J Inorg Biochem. 2019 Jun;195:61-70. doi: 10.1016/j.jinorgbio.2019.03.009. Epub 2019 Mar 21. J Inorg Biochem. 2019. PMID: 30925402 Free PMC article.
-
Understanding molecular enzymology of porphyrin-binding α + β barrel proteins - One fold, multiple functions.Biochim Biophys Acta Proteins Proteom. 2021 Jan;1869(1):140536. doi: 10.1016/j.bbapap.2020.140536. Epub 2020 Sep 4. Biochim Biophys Acta Proteins Proteom. 2021. PMID: 32891739 Free PMC article. Review.
-
Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.ACS Catal. 2020 May 15;10(10):5405-5418. doi: 10.1021/acscatal.0c00411. Epub 2020 Apr 9. ACS Catal. 2020. PMID: 32440366 Free PMC article.
-
Investigating the Roles of Listeria monocytogenes Peroxidases in Growth and Virulence.Microbiol Spectr. 2021 Sep 3;9(1):e0044021. doi: 10.1128/Spectrum.00440-21. Epub 2021 Jul 21. Microbiol Spectr. 2021. PMID: 34287055 Free PMC article.
-
Insights into the flexibility of the domain-linking loop in actinobacterial coproheme decarboxylase through structures and molecular dynamics simulations.Protein Sci. 2025 Feb;34(2):e70027. doi: 10.1002/pro.70027. Protein Sci. 2025. PMID: 39865384 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

