Negative Selection by Spiral Inertial Microfluidics Improves Viral Recovery and Sequencing from Blood
- PMID: 29536737
- PMCID: PMC6195311
- DOI: 10.1021/acs.analchem.7b05200
Negative Selection by Spiral Inertial Microfluidics Improves Viral Recovery and Sequencing from Blood
Abstract
In blood samples from patients with viral infection, it is often important to separate viral particles from human cells, for example, to minimize background in performing viral whole genome sequencing. Here, we present a microfluidic device that uses spiral inertial microfluidics with continuous circulation to separate host cells from viral particles and free nucleic acid. We demonstrate that this device effectively reduces white blood cells, red blood cells, and platelets from both whole blood and plasma samples with excellent recovery of viral nucleic acid. Furthermore, microfluidic separation leads to greater viral genome coverage and depth, highlighting an important application of this device in processing clinical samples for viral genome sequencing.
Conflict of interest statement
Conflict of Interest Disclosure
The authors have filed a patent application on the technology described here.
Figures

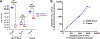
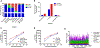
References
-
- McMullan LK; Folk SM; Kelly AJ; MacNeil A; Goldsmith CS; Metcalfe MG; Batten BC; Albariño CG; Zaki SR; Rollin PE; Nicholson WL; Nichol ST N. Engl. J. Med 2012, 367, 834–841. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

