How Tyramine β-Hydroxylase Controls the Production of Octopamine, Modulating the Mobility of Beetles
- PMID: 29538302
- PMCID: PMC5877707
- DOI: 10.3390/ijms19030846
How Tyramine β-Hydroxylase Controls the Production of Octopamine, Modulating the Mobility of Beetles
Abstract
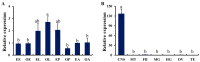
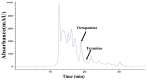
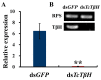
Biogenic amines perform many kinds of important physiological functions in the central nervous system (CNS) of insects, acting as neuromodulators, neurotransmitters, and neurohormones. The five most abundant types of biogenic amines in invertebrates are dopamine, histamine, serotonin, tyramine, and octopamine (OA). However, in beetles, an important group of model and pest insects, the role of tyramine β-hydroxylase (TβH) in the OA biosynthesis pathway and the regulation of behavior remains unknown so far. We therefore investigated the molecular characterization and spatiotemporal expression profiles of TβH in red flour beetles (Triboliun castaneum). Most importantly, we detected the production of OA and measured the crawling speed of beetles after dsTcTβH injection. We concluded that TcTβH controls the biosynthesis amount of OA in the CNS, and this in turn modulates the mobility of the beetles. Our new results provided basic information about the key genes in the OA biosynthesis pathway of the beetles, and expanded our knowledge on the physiological functions of OA in insects.
Keywords: Tribolium castaneum; mobility; octopamine; tyramine-β-hydroxylase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Octopamine is required for successful reproduction in the classical insect model, Rhodnius prolixus.PLoS One. 2024 Jul 12;19(7):e0306611. doi: 10.1371/journal.pone.0306611. eCollection 2024. PLoS One. 2024. PMID: 38995904 Free PMC article.
-
Altered Octopamine synthesis impairs tyrosine metabolism affecting Helicoverpa armigera vitality.Pestic Biochem Physiol. 2025 Mar;208:106323. doi: 10.1016/j.pestbp.2025.106323. Epub 2025 Feb 10. Pestic Biochem Physiol. 2025. PMID: 40015913
-
Characterization and developmental regulation of tyramine-beta-hydroxylase in the CNS of the moth, Manduca sexta.Insect Biochem Mol Biol. 2000 May;30(5):377-86. doi: 10.1016/s0965-1748(00)00011-4. Insect Biochem Mol Biol. 2000. PMID: 10745161
-
Molecular and pharmacological properties of insect biogenic amine receptors: lessons from Drosophila melanogaster and Apis mellifera.Arch Insect Biochem Physiol. 2001 Sep;48(1):13-38. doi: 10.1002/arch.1055. Arch Insect Biochem Physiol. 2001. PMID: 11519073 Review.
-
Tyramine and octopamine: ruling behavior and metabolism.Annu Rev Entomol. 2005;50:447-77. doi: 10.1146/annurev.ento.50.071803.130404. Annu Rev Entomol. 2005. PMID: 15355245 Review.
Cited by
-
Octopamine is required for successful reproduction in the classical insect model, Rhodnius prolixus.PLoS One. 2024 Jul 12;19(7):e0306611. doi: 10.1371/journal.pone.0306611. eCollection 2024. PLoS One. 2024. PMID: 38995904 Free PMC article.
-
Knockdown of a β-Adrenergic-Like Octopamine Receptor Affects Locomotion and Reproduction of Tribolium castaneum.Int J Mol Sci. 2021 Jul 6;22(14):7252. doi: 10.3390/ijms22147252. Int J Mol Sci. 2021. PMID: 34298876 Free PMC article.
-
The Role of Tyramine β-Hydroxylase in Density Dependent Immunityof Oriental Armyworm (Mythmina separata) Larva.Int J Mol Sci. 2019 Mar 28;20(7):1553. doi: 10.3390/ijms20071553. Int J Mol Sci. 2019. PMID: 30925699 Free PMC article.
References
-
- Chen A., Ng F., Lebestky T., Grygoruk A., Djapri C., Lawal H.O., Zaveri H.A., Mehanzel F., Najibi R., Seidman G., et al. Dispensable, redundant, complementary, and cooperative roles of dopamine, octopamine, and serotonin in Drosophila melanogaster. Genetics. 2013;193:159–176. doi: 10.1534/genetics.112.142042. - DOI - PMC - PubMed
-
- Burrell B.D., Smith B.H. Modulation of the honey bee (Apis Mellifera) sting response by octopamine. J. Insect Physiol. 1995;41:671–680. doi: 10.1016/0022-1910(95)00022-M. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

