Generalising better: Applying deep learning to integrate deleteriousness prediction scores for whole-exome SNV studies
- PMID: 29538399
- PMCID: PMC5851551
- DOI: 10.1371/journal.pone.0192829
Generalising better: Applying deep learning to integrate deleteriousness prediction scores for whole-exome SNV studies
Abstract
Many automatic classifiers were introduced to aid inference of phenotypical effects of uncategorised nsSNVs (nonsynonymous Single Nucleotide Variations) in theoretical and medical applications. Lately, several meta-estimators have been proposed that combine different predictors, such as PolyPhen and SIFT, to integrate more information in a single score. Although many advances have been made in feature design and machine learning algorithms used, the shortage of high-quality reference data along with the bias towards intensively studied in vitro models call for improved generalisation ability in order to further increase classification accuracy and handle records with insufficient data. Since a meta-estimator basically combines different scoring systems with highly complicated nonlinear relationships, we investigated how deep learning (supervised and unsupervised), which is particularly efficient at discovering hierarchies of features, can improve classification performance. While it is believed that one should only use deep learning for high-dimensional input spaces and other models (logistic regression, support vector machines, Bayesian classifiers, etc) for simpler inputs, we still believe that the ability of neural networks to discover intricate structure in highly heterogenous datasets can aid a meta-estimator. We compare the performance with various popular predictors, many of which are recommended by the American College of Medical Genetics and Genomics (ACMG), as well as available deep learning-based predictors. Thanks to hardware acceleration we were able to use a computationally expensive genetic algorithm to stochastically optimise hyper-parameters over many generations. Overfitting was hindered by noise injection and dropout, limiting coadaptation of hidden units. Although we stress that this work was not conceived as a tool comparison, but rather an exploration of the possibilities of deep learning application in ensemble scores, our results show that even relatively simple modern neural networks can significantly improve both prediction accuracy and coverage. We provide open-access to our finest model via the web-site: http://score.generesearch.ru/services/badmut/.
Conflict of interest statement
Figures
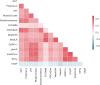
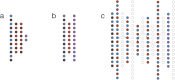

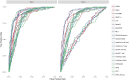
References
-
- Ng SB, Nickerson DA, Bamshad MJ, Shendure J. Massively parallel sequencing and rare disease. Human Molecular Genetics. 2010;19(R2):R119–R124. doi: 10.1093/hmg/ddq390 - DOI - PMC - PubMed
-
- Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Research. 2011;39(17):37–43. doi: 10.1093/nar/gkr407 - DOI - PMC - PubMed
-
- Ng PC, Henikoff S. Predicting the Effects of Amino Acid Substitutions on Protein Function. Annu Rev Genom Hum Genet. 2006;7(1):61–80. doi: 10.1146/annurev.genom.7.080505.115630 - DOI - PubMed
-
- Thusberg J, Vihinen M. Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Human Mutation. 2009;30(5):703–714. doi: 10.1002/humu.20938 - DOI - PubMed
-
- Cooper GM, Goode DL, Ng SB, Sidow A, Bamshad MJ, Shendure J, et al. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nature Methods. 2010;7(4):250–251. doi: 10.1038/nmeth0410-250 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

