Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis
- PMID: 29540203
- PMCID: PMC5853091
- DOI: 10.1186/s13059-018-1416-2
Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis
Abstract
Background: Organogenesis is crucial for proper organ formation during mammalian embryonic development. However, the similarities and shared features between different organs and the cellular heterogeneity during this process at single-cell resolution remain elusive.
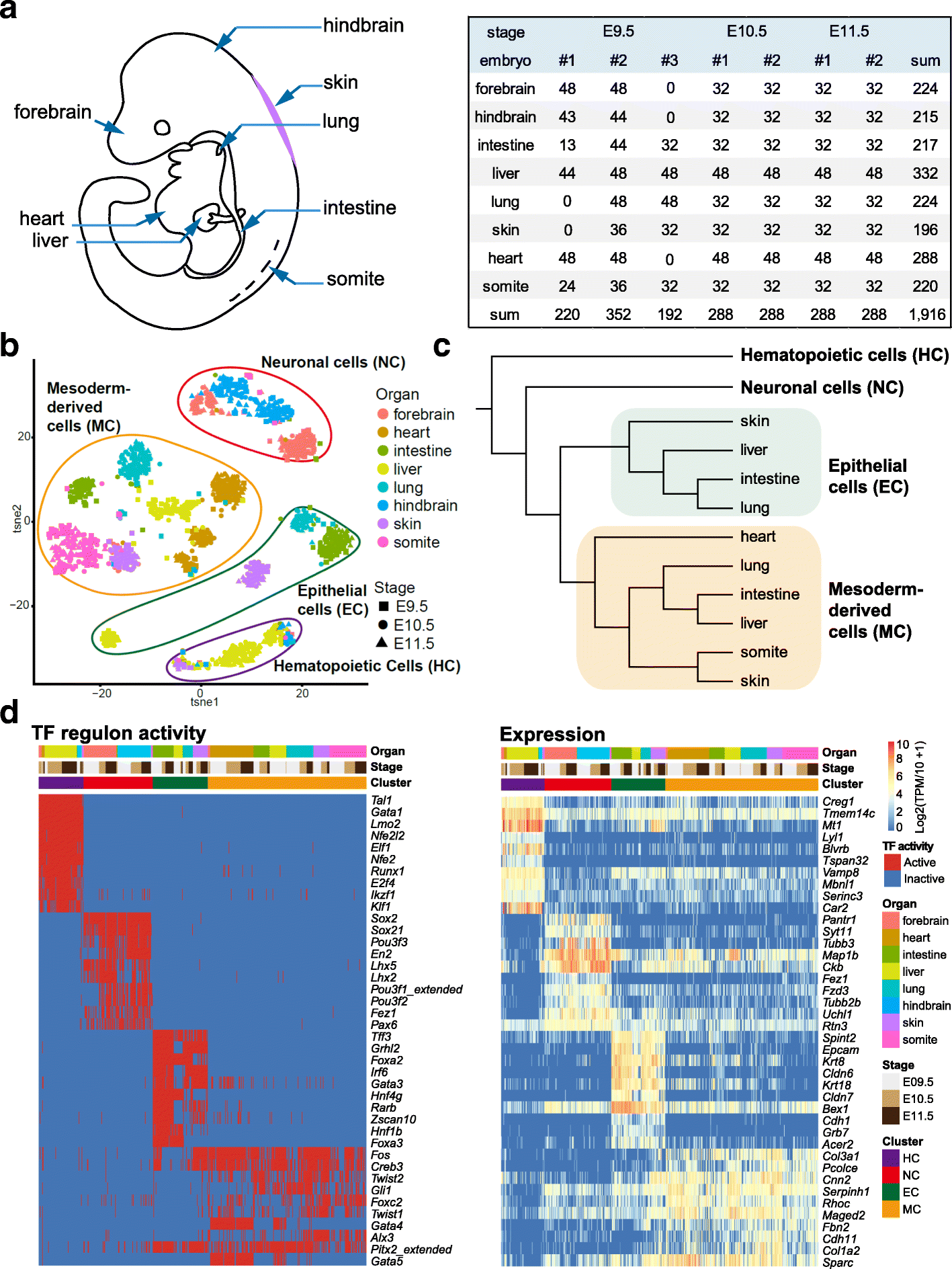
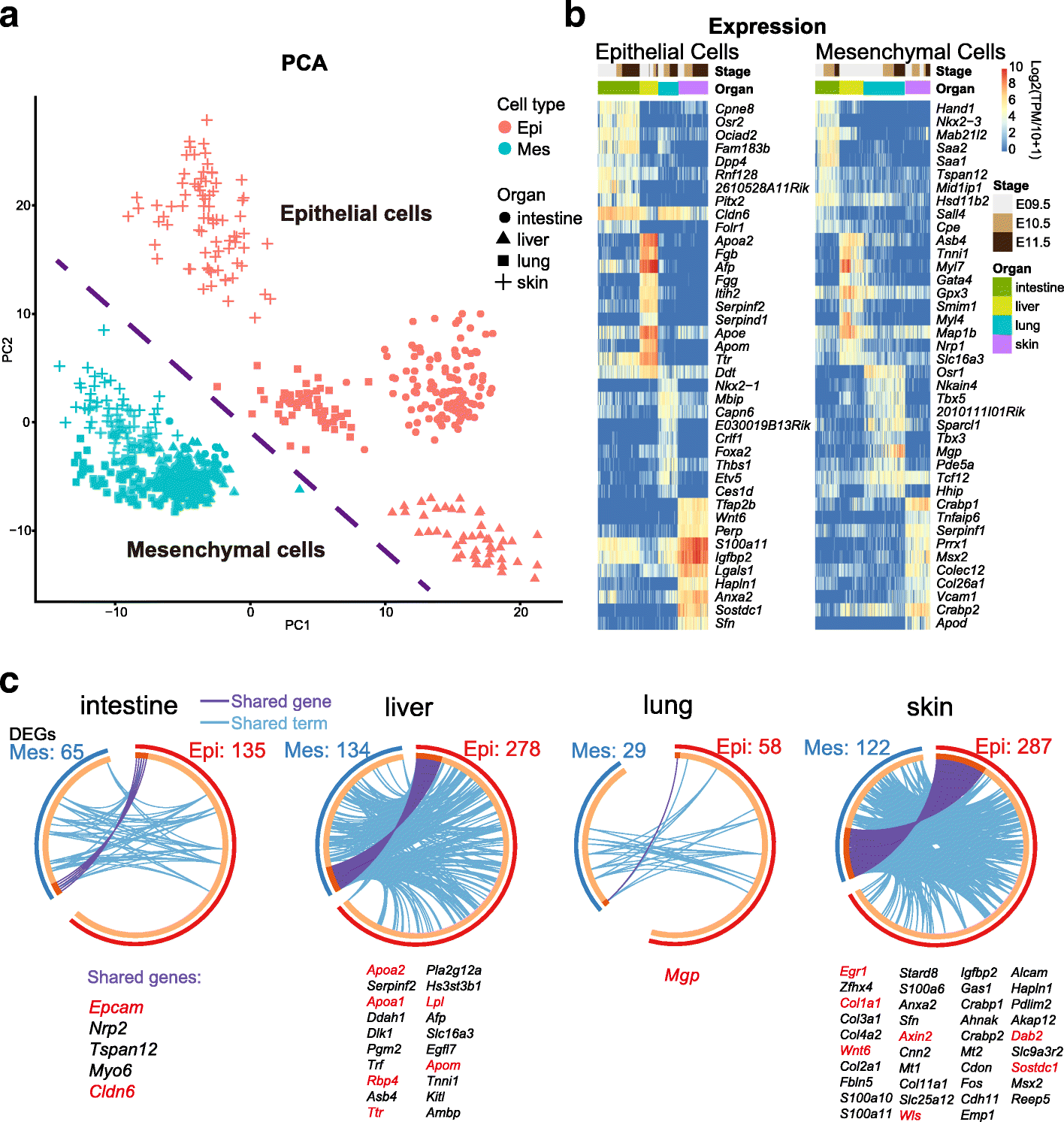
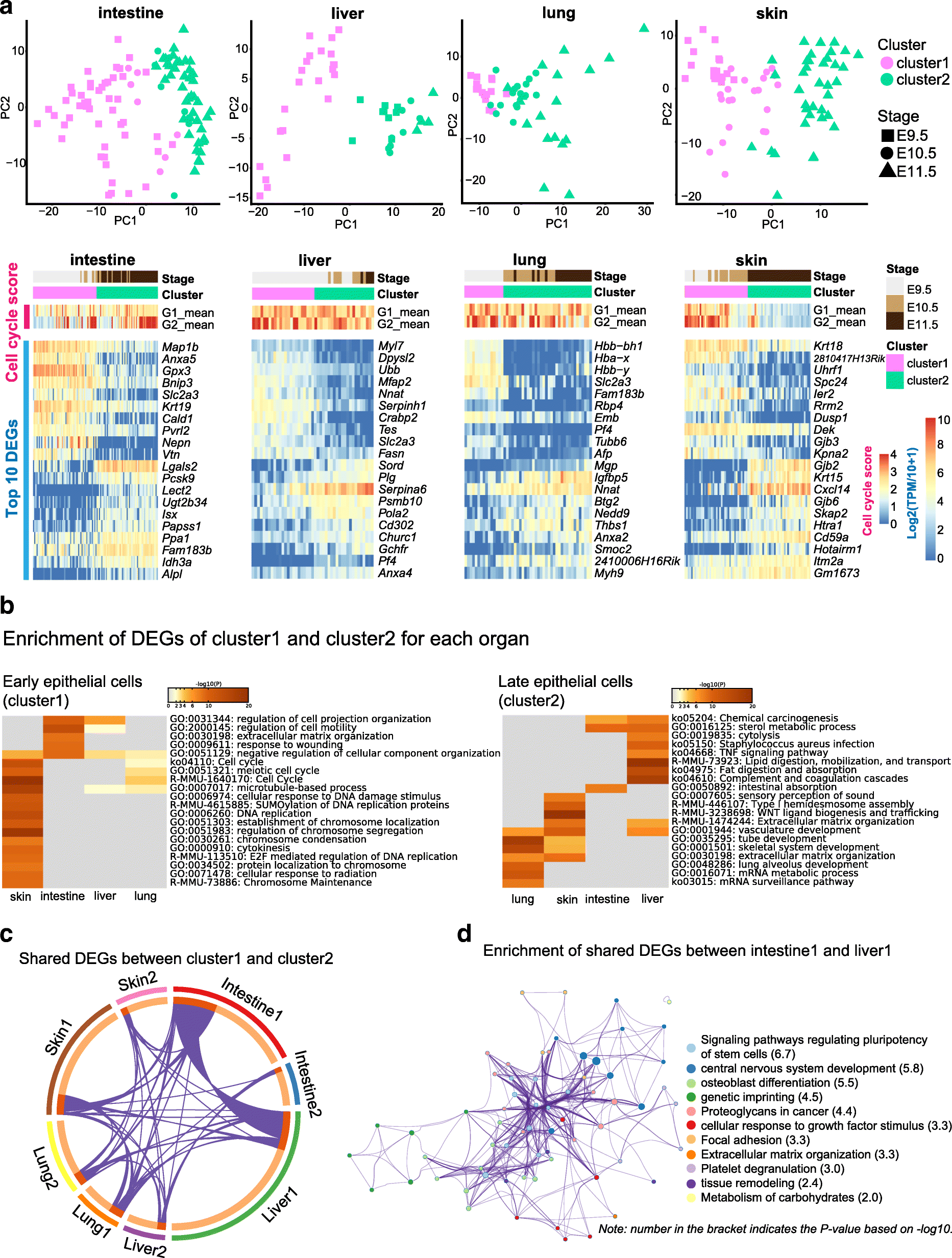
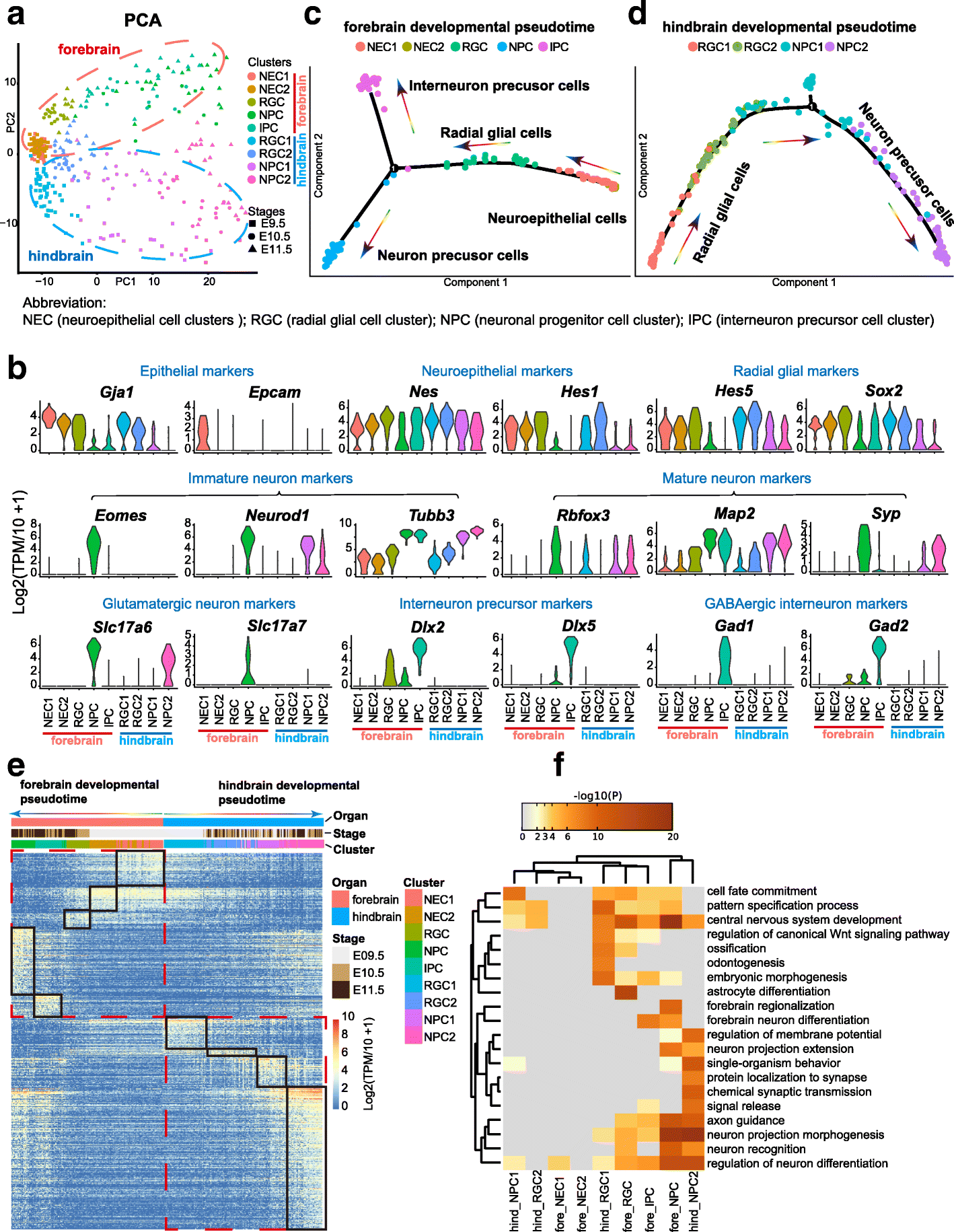
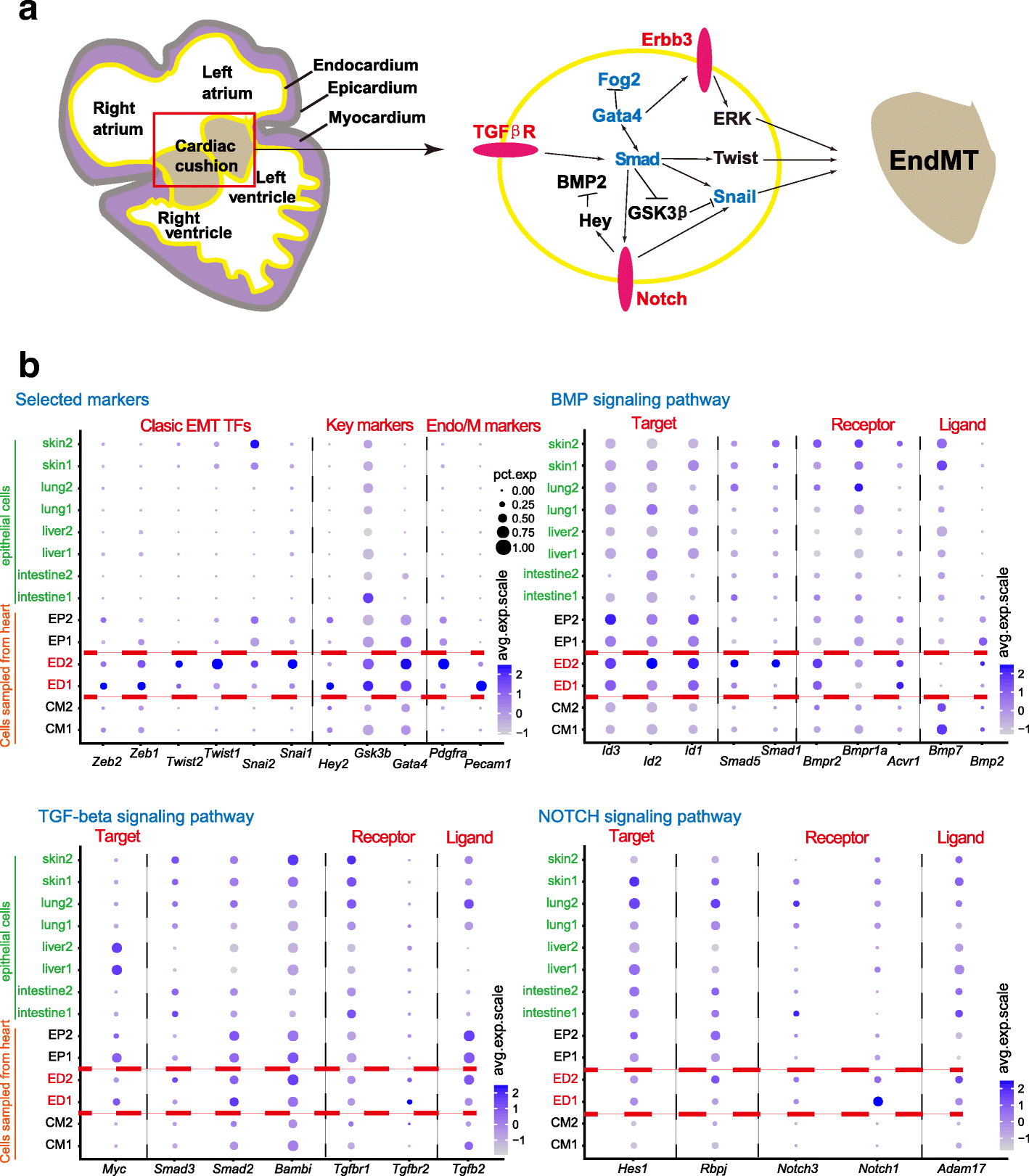
Results: We perform single-cell RNA sequencing analysis of 1916 individual cells from eight organs and tissues of E9.5 to E11.5 mouse embryos, namely, the forebrain, hindbrain, skin, heart, somite, lung, liver, and intestine. Based on the regulatory activities rather than the expression patterns, all cells analyzed can be well classified into four major groups with epithelial, mesodermal, hematopoietic, and neuronal identities. For different organs within the same group, the similarities and differences of their features and developmental paths are revealed and reconstructed.
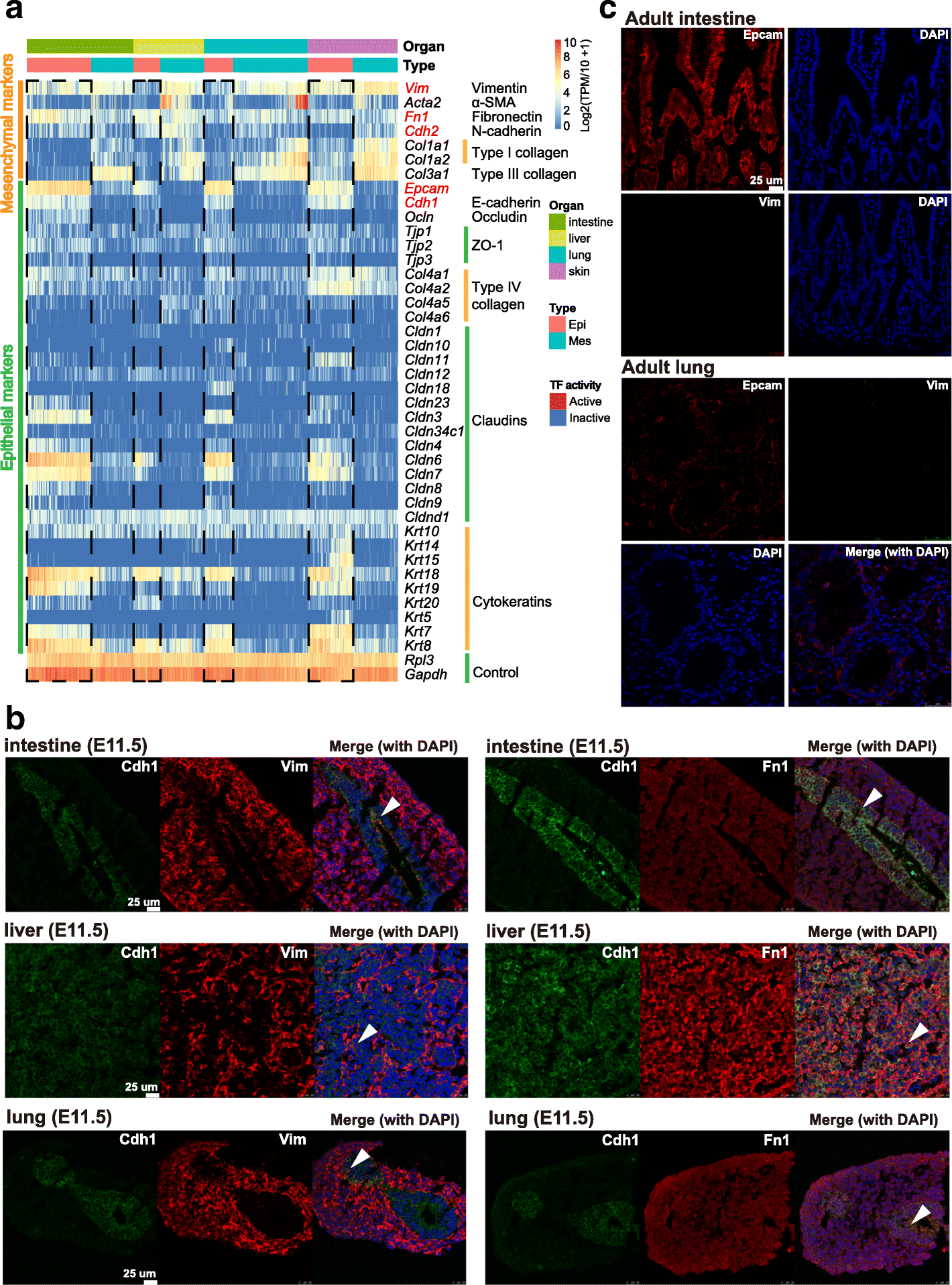
Conclusions: We identify mutual interactions between epithelial and mesenchymal cells and detect epithelial cells with prevalent mesenchymal features during organogenesis, which are similar to the features of intermediate epithelial/mesenchymal cells during tumorigenesis. The comprehensive transcriptome at single-cell resolution profiled in our study paves the way for future mechanistic studies of the gene-regulatory networks governing mammalian organogenesis.
Keywords: Epithelial/mesenchymal hybrid state; Interactions between mesenchyme and epithelium; Organogenesis; Single-cell RNA-seq.
Conflict of interest statement
Ethics approval
The study was approved by the Peking University Institutional Animal Care and Use Committee (IACUC). All the animal experiments were conducted following their guidelines.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
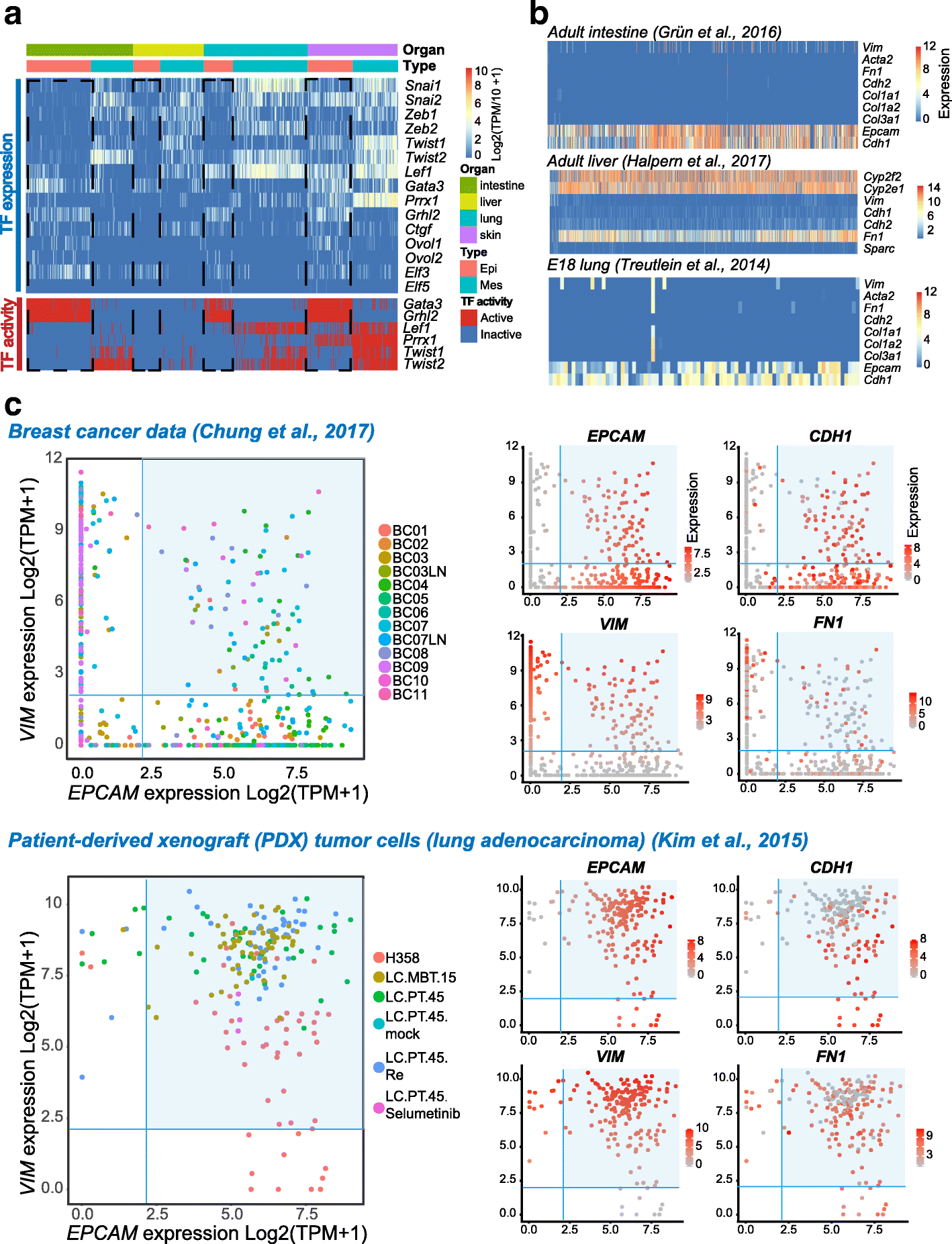
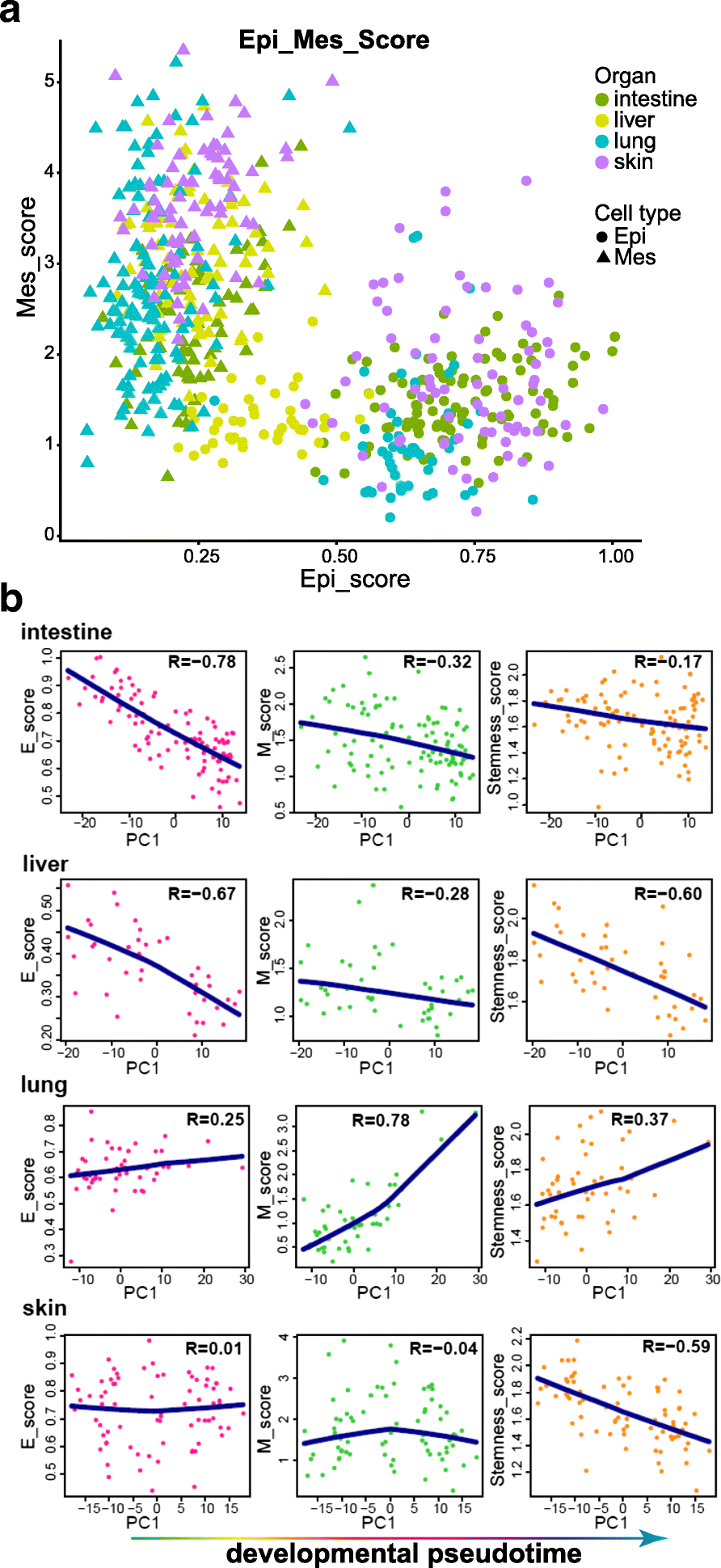
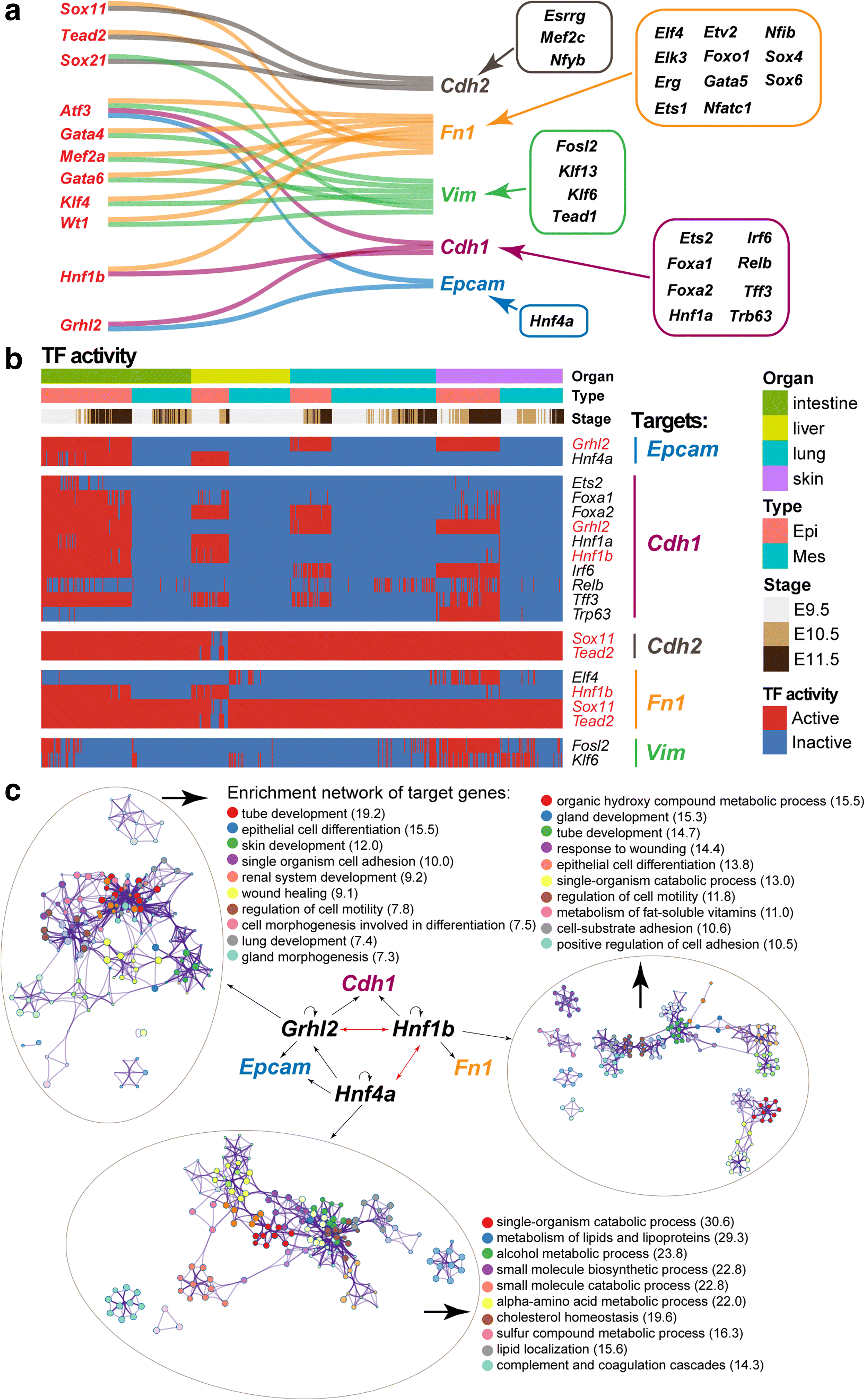
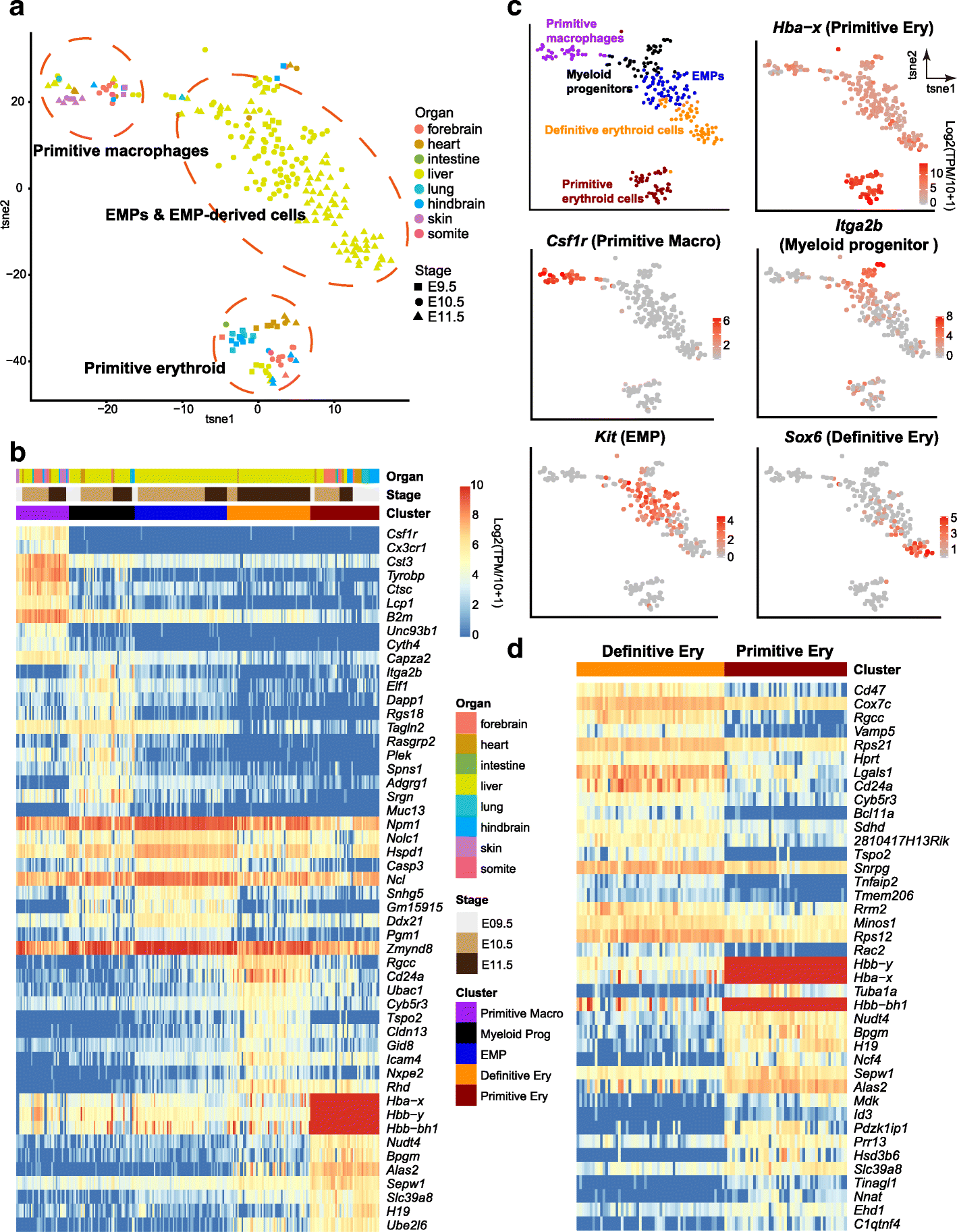
Similar articles
-
Identification of genes expressed in a mesenchymal subset regulating prostate organogenesis using tissue and single cell transcriptomics.Sci Rep. 2017 Nov 27;7(1):16385. doi: 10.1038/s41598-017-16685-8. Sci Rep. 2017. PMID: 29180763 Free PMC article.
-
The single-cell transcriptional landscape of mammalian organogenesis.Nature. 2019 Feb;566(7745):496-502. doi: 10.1038/s41586-019-0969-x. Epub 2019 Feb 20. Nature. 2019. PMID: 30787437 Free PMC article.
-
Single cell transcriptomics identifies a signaling network coordinating endoderm and mesoderm diversification during foregut organogenesis.Nat Commun. 2020 Aug 27;11(1):4158. doi: 10.1038/s41467-020-17968-x. Nat Commun. 2020. PMID: 32855417 Free PMC article.
-
Exosomal microRNA communication between tissues during organogenesis.RNA Biol. 2017 Dec 2;14(12):1683-1689. doi: 10.1080/15476286.2017.1361098. Epub 2017 Sep 29. RNA Biol. 2017. PMID: 28816640 Free PMC article. Review.
-
Epithelial-mesenchymal interactions: a fundamental Developmental Biology mechanism.Int J Dev Biol. 2014;58(5):303-6. doi: 10.1387/ijdb.140143dr. Int J Dev Biol. 2014. PMID: 25354449 Review.
Cited by
-
Single-cell profiling reveals molecular basis of malignant phenotypes and tumor microenvironments in small bowel adenocarcinomas.Cell Discov. 2022 Sep 14;8(1):92. doi: 10.1038/s41421-022-00434-x. Cell Discov. 2022. PMID: 36104333 Free PMC article.
-
Decoding the activated stem cell phenotype of the neonatally maturing pituitary.Elife. 2022 Jun 14;11:e75742. doi: 10.7554/eLife.75742. Elife. 2022. PMID: 35699412 Free PMC article.
-
Fetal liver development and implications for liver disease pathogenesis.Nat Rev Gastroenterol Hepatol. 2023 Sep;20(9):561-581. doi: 10.1038/s41575-023-00775-2. Epub 2023 May 19. Nat Rev Gastroenterol Hepatol. 2023. PMID: 37208503 Review.
-
A Refined Single Cell Landscape of Haematopoiesis in the Mouse Foetal Liver.J Dev Biol. 2023 Mar 23;11(2):15. doi: 10.3390/jdb11020015. J Dev Biol. 2023. PMID: 37092477 Free PMC article.
-
Targeting SMAD-Dependent Signaling: Considerations in Epithelial and Mesenchymal Solid Tumors.Pharmaceuticals (Basel). 2024 Mar 1;17(3):326. doi: 10.3390/ph17030326. Pharmaceuticals (Basel). 2024. PMID: 38543112 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

