An RNA-binding compound that stabilizes the HIV-1 gRNA packaging signal structure and specifically blocks HIV-1 RNA encapsidation
- PMID: 29540207
- PMCID: PMC5853050
- DOI: 10.1186/s12977-018-0407-4
An RNA-binding compound that stabilizes the HIV-1 gRNA packaging signal structure and specifically blocks HIV-1 RNA encapsidation
Abstract
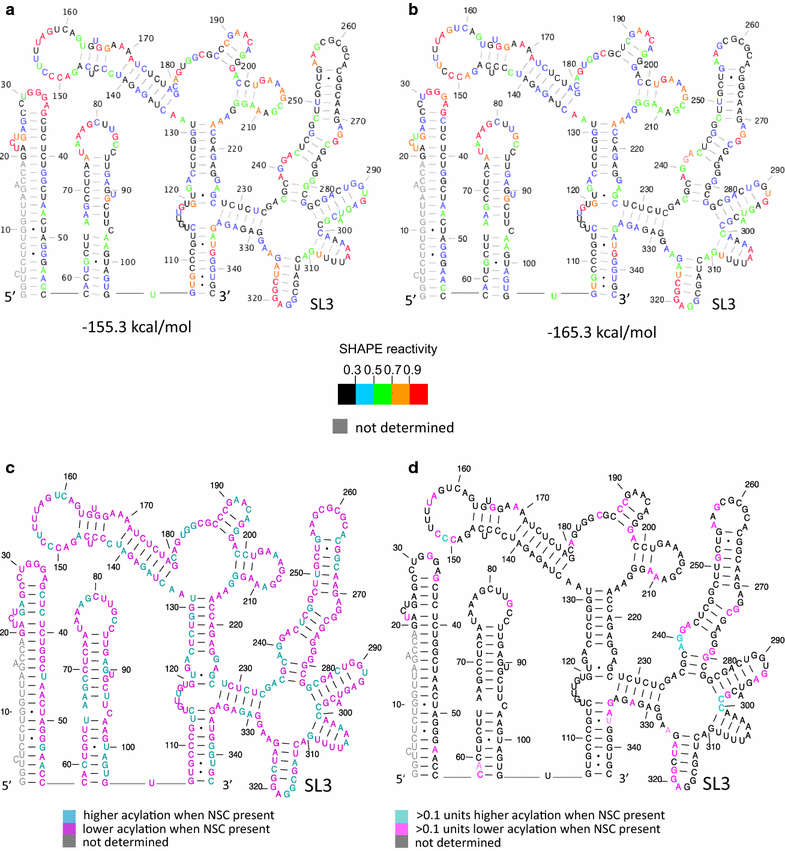
Background: NSC260594, a quinolinium derivative from the NCI diversity set II compound library, was previously identified in a target-based assay as an inhibitor of the interaction between the HIV-1 (ψ) stem-loop 3 (SL3) RNA and Gag. This compound was shown to exhibit potent antiviral activity. Here, the effects of this compound on individual stages of the viral lifecycle were examined by qRT-PCR, ELISA and Western blot, to see if its actions were specific to the viral packaging stage. The structural effects of NSC260594 binding to the HIV-1 gRNA were also examined by SHAPE and dimerization assays.
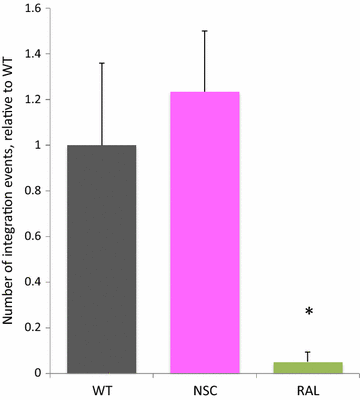
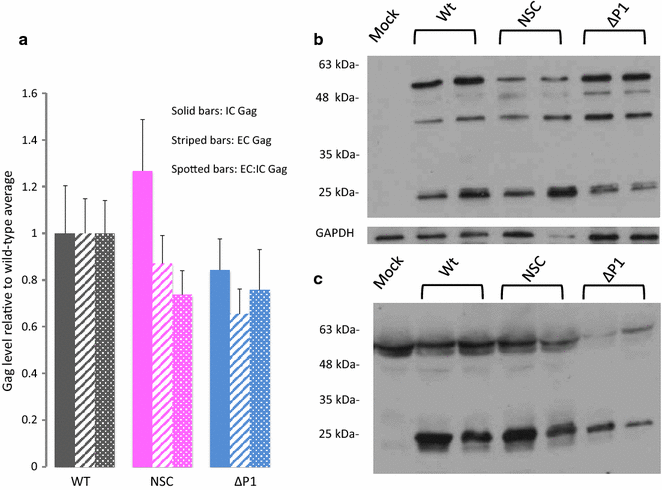
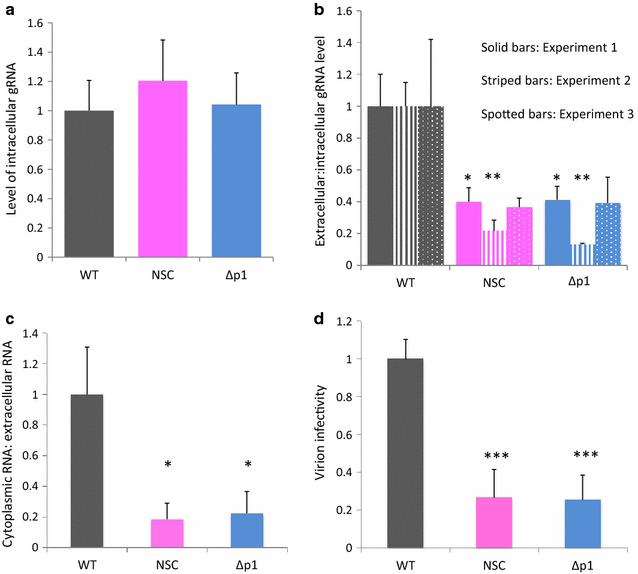
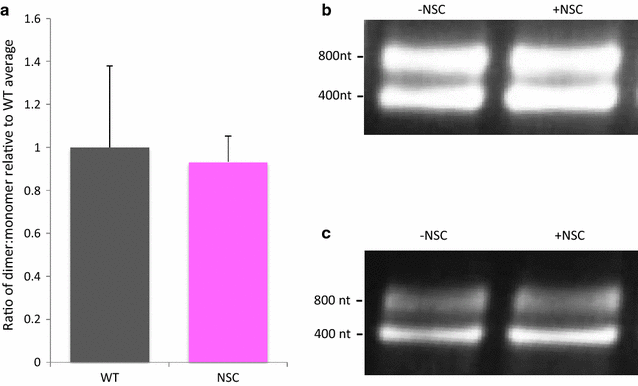
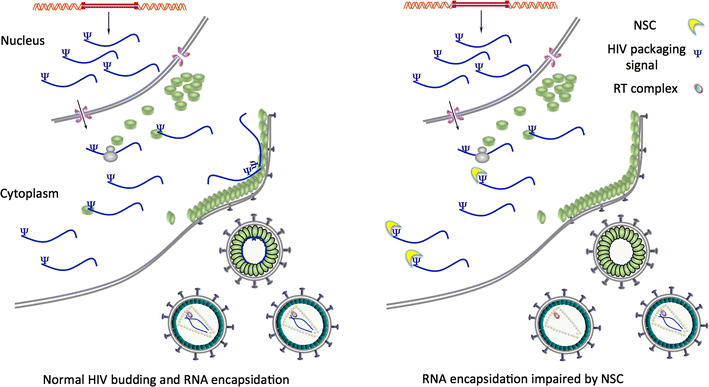
Results: Treatment of cells with NSC260594 did not reduce the number of integration events of incoming virus, and treatment of virus producing cells did not affect the level of intracellular Gag protein or viral particle release as determined by immunoblot. However, NSC260594 reduced the incorporation of gRNA into virions by up to 82%, without affecting levels of gRNA inside the cell. This reduction in packaging correlated closely with the reduction in infectivity of the released viral particles. To establish the structural effects of NSC260594 on the HIV-1 gRNA, we performed SHAPE analyses to pinpoint RNA structural changes. NSC260594 had a stabilizing effect on the wild type RNA that was not confined to SL3, but that was propagated across the structure. A packaging mutant lacking SL3 did not show this effect.
Conclusions: NSC260594 acts as a specific inhibitor of HIV-1 RNA packaging. No other viral functions are affected. Its action involves preventing the interaction of Gag with SL3 by stabilizing this small RNA stem-loop which then leads to stabilization of the global packaging signal region (psi or ψ). This confirms data, previously only shown in analyses of isolated SL3 oligonucleotides, that SL3 is structurally labile in the presence of Gag and that this is critical for the complete psi region to be able to adopt different conformations. Since replication is otherwise unaffected by NSC260594 the flexibility of SL3 appears to be a unique requirement for genome encapsidation and identifies this process as a highly specific drug target. This study is proof of principle that development of a new class of antiretroviral drugs that specifically target viral packaging by binding to the viral genomic RNA is achievable.
Keywords: Antiretroviral drugs; HIV-1; Packaging; RNA structure.
Figures
Similar articles
-
HIV-1 integrase binding to genomic RNA 5'-UTR induces local structural changes in vitro and in virio.Retrovirology. 2021 Nov 22;18(1):37. doi: 10.1186/s12977-021-00582-0. Retrovirology. 2021. PMID: 34809662 Free PMC article.
-
Stabilizing role of structural elements within the 5´ Untranslated Region (UTR) and gag sequences in Mason-Pfizer monkey virus (MPMV) genomic RNA packaging.RNA Biol. 2019 May;16(5):612-625. doi: 10.1080/15476286.2019.1572424. Epub 2019 Feb 17. RNA Biol. 2019. PMID: 30773097 Free PMC article.
-
NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition.J Mol Biol. 2000 Aug 11;301(2):491-511. doi: 10.1006/jmbi.2000.3979. J Mol Biol. 2000. PMID: 10926523
-
On the Selective Packaging of Genomic RNA by HIV-1.Viruses. 2016 Sep 12;8(9):246. doi: 10.3390/v8090246. Viruses. 2016. PMID: 27626441 Free PMC article. Review.
-
Retroviral Gag protein-RNA interactions: Implications for specific genomic RNA packaging and virion assembly.Semin Cell Dev Biol. 2019 Feb;86:129-139. doi: 10.1016/j.semcdb.2018.03.015. Epub 2018 Apr 1. Semin Cell Dev Biol. 2019. PMID: 29580971 Free PMC article. Review.
Cited by
-
Coronavirus genomic RNA packaging.Virology. 2019 Nov;537:198-207. doi: 10.1016/j.virol.2019.08.031. Epub 2019 Aug 30. Virology. 2019. PMID: 31505321 Free PMC article. Review.
-
A purine loop and the primer binding site are critical for the selective encapsidation of mouse mammary tumor virus genomic RNA by Pr77Gag.Nucleic Acids Res. 2021 May 7;49(8):4668-4688. doi: 10.1093/nar/gkab223. Nucleic Acids Res. 2021. PMID: 33836091 Free PMC article.
-
Small Molecules Targeting Viral RNA.Int J Mol Sci. 2023 Aug 31;24(17):13500. doi: 10.3390/ijms241713500. Int J Mol Sci. 2023. PMID: 37686306 Free PMC article. Review.
-
HIV-1 Packaging Visualised by In-Gel SHAPE.Viruses. 2021 Nov 29;13(12):2389. doi: 10.3390/v13122389. Viruses. 2021. PMID: 34960658 Free PMC article.
-
Optimization of a lentivirus-mediated gene therapy targeting HIV-1 RNA to eliminate HIV-1-infected cells.Mol Ther Nucleic Acids. 2024 Sep 16;35(4):102341. doi: 10.1016/j.omtn.2024.102341. eCollection 2024 Dec 10. Mol Ther Nucleic Acids. 2024. PMID: 39434850 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

