Antibiotic Hybrids: the Next Generation of Agents and Adjuvants against Gram-Negative Pathogens?
- PMID: 29540434
- PMCID: PMC5967690
- DOI: 10.1128/CMR.00077-17
Antibiotic Hybrids: the Next Generation of Agents and Adjuvants against Gram-Negative Pathogens?
Abstract
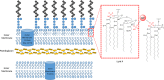
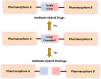
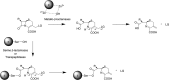
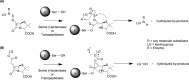
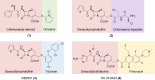
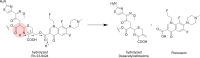
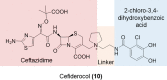
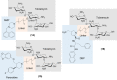
The global incidence of drug-resistant Gram-negative bacillary infections has been increasing, and there is a dire need to develop novel strategies to overcome this problem. Intrinsic resistance in Gram-negative bacteria, such as their protective outer membrane and constitutively overexpressed efflux pumps, is a major survival weapon that renders them refractory to current antibiotics. Several potential avenues to overcome this problem have been at the heart of antibiotic drug discovery in the past few decades. We review some of these strategies, with emphasis on antibiotic hybrids either as stand-alone antibacterial agents or as adjuvants that potentiate a primary antibiotic in Gram-negative bacteria. Antibiotic hybrid is defined in this review as a synthetic construct of two or more pharmacophores belonging to an established agent known to elicit a desired antimicrobial effect. The concepts, advances, and challenges of antibiotic hybrids are elaborated in this article. Moreover, we discuss several antibiotic hybrids that were or are in clinical evaluation. Mechanistic insights into how tobramycin-based antibiotic hybrids are able to potentiate legacy antibiotics in multidrug-resistant Gram-negative bacilli are also highlighted. Antibiotic hybrids indeed have a promising future as a therapeutic strategy to overcome drug resistance in Gram-negative pathogens and/or expand the usefulness of our current antibiotic arsenal.
Keywords: antibacterial; antibiotic; antimicrobial; efflux; hybrid; permeability.
Copyright © 2018 American Society for Microbiology.
Figures








Similar articles
-
Next-generation strategy for treating drug resistant bacteria: Antibiotic hybrids.Indian J Med Res. 2019 Feb;149(2):97-106. doi: 10.4103/ijmr.IJMR_755_18. Indian J Med Res. 2019. PMID: 31219074 Free PMC article. Review.
-
Antibiotic adjuvants: an alternative approach to overcome multi-drug resistant Gram-negative bacteria.Crit Rev Microbiol. 2019 May;45(3):301-314. doi: 10.1080/1040841X.2019.1599813. Epub 2019 Apr 15. Crit Rev Microbiol. 2019. PMID: 30985240 Review.
-
Amphiphilic nebramine-based hybrids Rescue legacy antibiotics from intrinsic resistance in multidrug-resistant Gram-negative bacilli.Eur J Med Chem. 2019 Aug 1;175:187-200. doi: 10.1016/j.ejmech.2019.05.003. Epub 2019 May 2. Eur J Med Chem. 2019. PMID: 31078866
-
Antibiotic adjuvants against multidrug-resistant Gram-negative bacteria: important component of future antimicrobial therapy.Microbiol Res. 2024 Oct;287:127842. doi: 10.1016/j.micres.2024.127842. Epub 2024 Jul 18. Microbiol Res. 2024. PMID: 39032266 Review.
-
Multidrug-resistant gram-negative organisms: a review of recently approved antibiotics and novel pipeline agents.Int J Clin Pharm. 2020 Aug;42(4):1016-1025. doi: 10.1007/s11096-020-01089-y. Epub 2020 Jul 7. Int J Clin Pharm. 2020. PMID: 32638294 Review.
Cited by
-
Phage Therapy for Mycobacterium Abscessus and Strategies to Improve Outcomes.Microorganisms. 2021 Mar 14;9(3):596. doi: 10.3390/microorganisms9030596. Microorganisms. 2021. PMID: 33799414 Free PMC article. Review.
-
Novel Strategy to Combat Antibiotic Resistance: A Sight into the Combination of CRISPR/Cas9 and Nanoparticles.Pharmaceutics. 2021 Mar 8;13(3):352. doi: 10.3390/pharmaceutics13030352. Pharmaceutics. 2021. PMID: 33800235 Free PMC article. Review.
-
IMT-P8 potentiates Gram-positive specific antibiotics in intrinsically resistant Gram-negative bacteria.Antimicrob Agents Chemother. 2024 Oct 8;68(10):e0075324. doi: 10.1128/aac.00753-24. Epub 2024 Sep 5. Antimicrob Agents Chemother. 2024. PMID: 39235250 Free PMC article.
-
Facile synthesis, antimicrobial activity, and molecular docking analysis of 8-hydroxyquinoline-4-thiazolidinone hybrids.Future Med Chem. 2025 Feb;17(4):435-447. doi: 10.1080/17568919.2025.2463876. Epub 2025 Feb 14. Future Med Chem. 2025. PMID: 39949271
-
Repurposed Antimicrobial Combination Therapy: Tobramycin-Ciprofloxacin Hybrid Augments Activity of the Anticancer Drug Mitomycin C Against Multidrug-Resistant Gram-Negative Bacteria.Front Microbiol. 2019 Jul 10;10:1556. doi: 10.3389/fmicb.2019.01556. eCollection 2019. Front Microbiol. 2019. PMID: 31354660 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

