Structural and biochemical characterization of Siw14: A protein-tyrosine phosphatase fold that metabolizes inositol pyrophosphates
- PMID: 29540476
- PMCID: PMC5936820
- DOI: 10.1074/jbc.RA117.001670
Structural and biochemical characterization of Siw14: A protein-tyrosine phosphatase fold that metabolizes inositol pyrophosphates
Abstract
Inositol pyrophosphates (PP-InsPs) are "energetic" intracellular signals that are ubiquitous in animals, plants, and fungi; structural and biochemical characterization of PP-InsP metabolic enzymes provides insight into their evolution, reaction mechanisms, and regulation. Here, we describe the 2.35-Å-resolution structure of the catalytic core of Siw14, a 5-PP-InsP phosphatase from Saccharomyces cerevisiae and a member of the protein tyrosine-phosphatase (PTP) superfamily. Conclusions that we derive from structural data are supported by extensive site-directed mutagenesis and kinetic analyses, thereby attributing new functional significance to several key residues. We demonstrate the high activity and exquisite specificity of Siw14 for the 5-diphosphate group of PP-InsPs. The three structural elements that demarcate a 9.2-Å-deep substrate-binding pocket each have spatial equivalents in PTPs, but we identify how these are specialized for Siw14 to bind and hydrolyze the intensely negatively charged PP-InsPs. (a) The catalytic P-loop with the CX5R(S/T) PTP motif contains additional, positively charged residues. (b) A loop between the α5 and α6 helices, corresponding to the Q-loop in PTPs, contains a lysine and an arginine that extend into the catalytic pocket due to displacement of the α5 helix orientation through intramolecular crowding caused by three bulky, hydrophobic residues. (c) The general-acid loop in PTPs is replaced in Siw14 with a flexible loop that does not use an aspartate or glutamate as a general acid. We propose that an acidic residue is not required for phosphoanhydride hydrolysis.
Keywords: crystal structure; dual-specificity phosphoprotein phosphatase; enzyme mechanism; inositol phosphate; phosphatase.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

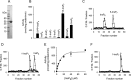
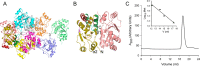
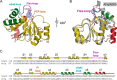
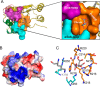
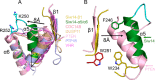
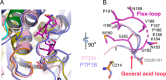
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

