Enhancer identification and activity evaluation in the red flour beetle, Tribolium castaneum
- PMID: 29540499
- PMCID: PMC11736658
- DOI: 10.1242/dev.160663
Enhancer identification and activity evaluation in the red flour beetle, Tribolium castaneum
Abstract
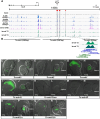
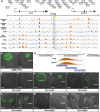
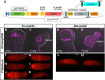
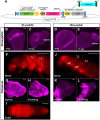
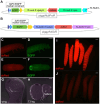
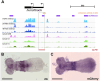
Evolution of cis-regulatory elements (such as enhancers) plays an important role in the production of diverse morphology. However, a mechanistic understanding is often limited by the absence of methods for studying enhancers in species other than established model systems. Here, we sought to establish methods to identify and test enhancer activity in the red flour beetle, Tribolium castaneum To identify possible enhancer regions, we first obtained genome-wide chromatin profiles from various tissues and stages of Tribolium using FAIRE (formaldehyde-assisted isolation of regulatory elements)-sequencing. Comparison of these profiles revealed a distinct set of open chromatin regions in each tissue and at each stage. In addition, comparison of the FAIRE data with sets of computationally predicted (i.e. supervised cis-regulatory module-predicted) enhancers revealed a very high overlap between the two datasets. Second, using nubbin in the wing and hunchback in the embryo as case studies, we established the first universal reporter assay system that works in various contexts in Tribolium, and in a cross-species context. Together, these advances will facilitate investigation of cis-evolution and morphological diversity in Tribolium and other insects.
Keywords: Chromatin profiling; FAIRE-seq; Insects; Reporter assay.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

