Positive Regulation of Hepatitis E Virus Replication by MicroRNA-122
- PMID: 29540601
- PMCID: PMC5952168
- DOI: 10.1128/JVI.01999-17
Positive Regulation of Hepatitis E Virus Replication by MicroRNA-122
Abstract
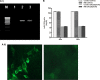
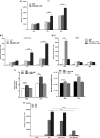
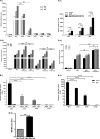
The molecular mechanisms of liver pathology and clinical disease in hepatitis E virus (HEV) infection remain unclear. MicroRNAs (miRNAs) are known to modulate viral pathogenesis either by directly altering viral gene expression or by enhancing cellular antiviral responses. Given the importance of microRNA-122 (miR-122) in liver pathobiology, we investigated possible role of miR-122 in HEV infection. In silico predictions using HEV genotype 1 (HEV-1), HEV-2, HEV-3, and HEV-4 sequences showed that the majority of genomes (203/222) harbor at least one miR-122/microRNA-122-3p (miR-122*) target site. Interestingly, HEV-1 genomes showed a highly (97%) conserved miR-122 target site in the RNA-dependent RNA polymerase (RdRp) region (RdRpc). We analyzed the significance of miR-122 target sites in HEV-1/HEV-3 (HEV-1/3) genomes by using a replicon-based cell culture system. HEV infection did not change the basal levels of miR-122 in hepatoma cells. However, transfection of these cells with miR-122 mimics enhanced HEV-1/3 replication and depletion of miR-122 with inhibitors led to suppression of HEV-1/3 replication. Mutant HEV-1 replicons with an altered target RdRpc sequence (CACTCC) showed a drastic decrease in virus replication, whereas introduction of alternative miR-122 target sites in mutant replicons rescued viral replication. There was enrichment of HEV-1 RNA and miR-122 molecules in RNA-induced silencing complexes in HEV-infected cells. Furthermore, pulldown of miR-122 molecules from HEV-infected cells resulted in pulldown of HEV genomic RNA along with miR-122 molecules. These observations indicate that miR-122 facilitates HEV-1 replication, probably via direct interaction with a target site in the viral genome. The positive role of miR-122 in viral replication presents novel opportunities for antiviral therapy and management of hepatitis E.IMPORTANCE Hepatitis E is a problem in both developing and developed countries. HEV infection in most patients follows a self-limited course; however, 20% to 30% mortality is seen in infected pregnant women. HEV superinfections in patients with chronic hepatitis B or hepatitis C virus infections are associated with adverse clinical outcomes, and both conditions warrant therapy. Chronic HEV infections in immunocompromised transplant recipients are known to rapidly progress into cirrhosis. Currently, off-label use of ribavirin (RBV) and polyethylene glycol-interferon (PEG-IFN) as antiviral therapy has shown promising results in both acute and chronic hepatitis E patients; however, the teratogenicity of RBV limits its use during pregnancy, while alpha IFN (IFN-α) increases the risk of transplant rejections. Experimental data determined with genotype 1 virus in the current study show that miR-122 facilitates HEV replication. These observations present novel opportunities for antiviral therapy and management of hepatitis E.
Keywords: Ago2; hepatitis E virus; interaction; miR-122; target site; virus replication.
Copyright © 2018 American Society for Microbiology.
Figures




Similar articles
-
ISG15 Modulates Type I Interferon Signaling and the Antiviral Response during Hepatitis E Virus Replication.J Virol. 2017 Sep 12;91(19):e00621-17. doi: 10.1128/JVI.00621-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28724761 Free PMC article.
-
Ribavirin Treatment Failure-Associated Mutation, Y1320H, in the RNA-Dependent RNA Polymerase of Genotype 3 Hepatitis E Virus (HEV) Enhances Virus Replication in a Rabbit HEV Infection Model.mBio. 2023 Apr 25;14(2):e0337222. doi: 10.1128/mbio.03372-22. Epub 2023 Feb 21. mBio. 2023. PMID: 36809085 Free PMC article.
-
Zinc Salts Block Hepatitis E Virus Replication by Inhibiting the Activity of Viral RNA-Dependent RNA Polymerase.J Virol. 2017 Oct 13;91(21):e00754-17. doi: 10.1128/JVI.00754-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28814517 Free PMC article.
-
Advances in Hepatitis E Virus Biology and Pathogenesis.Viruses. 2021 Feb 9;13(2):267. doi: 10.3390/v13020267. Viruses. 2021. PMID: 33572257 Free PMC article. Review.
-
Zinc: A Potential Antiviral Against Hepatitis E Virus Infection?DNA Cell Biol. 2018 Jul;37(7):593-599. doi: 10.1089/dna.2018.4175. Epub 2018 Jun 13. DNA Cell Biol. 2018. PMID: 29897788 Review.
Cited by
-
Structural aspects of hepatitis E virus.Arch Virol. 2022 Dec;167(12):2457-2481. doi: 10.1007/s00705-022-05575-8. Epub 2022 Sep 13. Arch Virol. 2022. PMID: 36098802 Free PMC article. Review.
-
Hepatitis E Virus Immunopathogenesis.Pathogens. 2021 Sep 13;10(9):1180. doi: 10.3390/pathogens10091180. Pathogens. 2021. PMID: 34578211 Free PMC article. Review.
-
Expression Profiles of Exosomal MicroRNAs from HEV- and HCV-Infected Blood Donors and Patients: A Pilot Study.Viruses. 2020 Jul 30;12(8):833. doi: 10.3390/v12080833. Viruses. 2020. PMID: 32751663 Free PMC article.
-
The Roles of Epinephelus coioides miR-122 in SGIV Infection and Replication.Mar Biotechnol (NY). 2021 Apr;23(2):294-307. doi: 10.1007/s10126-021-10023-w. Epub 2021 Feb 11. Mar Biotechnol (NY). 2021. PMID: 33570690 Free PMC article.
-
Differential miRNA signatures in Hepatitis E Virus Infection: Insights into acute, chronic, and pregnancy-related outcomes.Eur J Clin Microbiol Infect Dis. 2025 Jul 14. doi: 10.1007/s10096-025-05207-4. Online ahead of print. Eur J Clin Microbiol Infect Dis. 2025. PMID: 40660006 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

