Expression of lncRNA MIR222HG co-transcribed from the miR-221/222 gene promoter facilitates the development of castration-resistant prostate cancer
- PMID: 29540675
- PMCID: PMC5852960
- DOI: 10.1038/s41389-018-0039-5
Expression of lncRNA MIR222HG co-transcribed from the miR-221/222 gene promoter facilitates the development of castration-resistant prostate cancer
Abstract
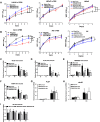
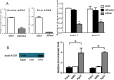
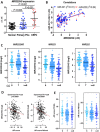
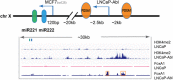
Mechanisms by which non-coding RNAs contribute to the progression of hormone-sensitive prostate cancer (PCa) (HSPC) to castration-resistant PCa (CRPC) remain largely unknown. We previously showed that microRNA-221/222 is up-regulated in CRPC and plays a critical role in modulating androgen receptor function during CRPC development. With further investigation, we characterized a putative promoter region located 23.3 kb upstream of the miR-221/222 gene, and this promoter is differentially activated in CRPC LNCaP-Abl cells, leading to the up-regulation of miR-221/222. Upon promoter activation, a set of polyadenylated long non-coding RNA (lncRNA) MIR222HGs was transcribed from this promoter region. Over-expression of these MIR222HGs increased androgen-independent cell growth and repressed the expression of androgen receptor-regulated dihydrotestosterone (DHT)-induced KLK3, TMPRSS2, and FKBP5 in HSPC LNCaP cells, hallmarks of the CRPC phenotype. Clinically, increased expression of MIR222HG is associated with PCa progression to CRPC. In primary tumors, expression levels of MIR222HG and miR-221/222 inversely correlate with Gleason score and androgen receptor (AR) pathway activity. Interestingly, MIR222HG is Argonaute 2-bound and its expression is Dicer 1-dependent, suggesting its functional association with the RNA-induced silencing complex. Further studies led to the hypothesis that MIR222HG may potentially affect miR-mediated expression silencing, subsequently leading to AR reprogramming. Our study highlights an essential role of a non-coding RNA in CRPC development and that differential activation of a single promoter can up-regulate two different types of non-coding RNAs, miR-221/222 and lncRNA MIR222HG, in CRPC. Additionally, this study reveals a novel function of lncRNAs as a modulator of Argonaute-mediated RNA-induced silencing complex.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Transcript Levels of Androgen Receptor Variant 7 and Ubiquitin-Conjugating Enzyme 2C in Hormone Sensitive Prostate Cancer and Castration-Resistant Prostate Cancer.Prostate. 2017 Jan;77(1):60-71. doi: 10.1002/pros.23248. Epub 2016 Aug 22. Prostate. 2017. PMID: 27550197
-
Estrogen induces androgen-repressed SOX4 expression to promote progression of prostate cancer cells.Prostate. 2015 Sep;75(13):1363-75. doi: 10.1002/pros.23017. Epub 2015 May 27. Prostate. 2015. PMID: 26015225
-
A novel AR translational regulator lncRNA LBCS inhibits castration resistance of prostate cancer.Mol Cancer. 2019 Jun 20;18(1):109. doi: 10.1186/s12943-019-1037-8. Mol Cancer. 2019. PMID: 31221168 Free PMC article.
-
Androgen receptors in hormone-dependent and castration-resistant prostate cancer.Pharmacol Ther. 2013 Dec;140(3):223-38. doi: 10.1016/j.pharmthera.2013.07.003. Epub 2013 Jul 13. Pharmacol Ther. 2013. PMID: 23859952 Review.
-
Non-Coding RNAs in Castration-Resistant Prostate Cancer: Regulation of Androgen Receptor Signaling and Cancer Metabolism.Int J Mol Sci. 2015 Dec 4;16(12):28943-78. doi: 10.3390/ijms161226138. Int J Mol Sci. 2015. PMID: 26690121 Free PMC article. Review.
Cited by
-
SPI1-mediated MIR222HG transcription promotes proneural-to-mesenchymal transition of glioma stem cells and immunosuppressive polarization of macrophages.Theranostics. 2023 May 27;13(10):3310-3329. doi: 10.7150/thno.82590. eCollection 2023. Theranostics. 2023. PMID: 37351164 Free PMC article.
-
Integrated analysis of lncRNA, miRNA and mRNA profiles reveals potential lncRNA functions during early HIV infection.J Transl Med. 2021 Apr 1;19(1):135. doi: 10.1186/s12967-021-02802-9. J Transl Med. 2021. PMID: 33794921 Free PMC article.
-
lncRNA FLVCR1-AS1 regulates cell proliferation, migration and invasion by sponging miR-485-5p in human cholangiocarcinoma.Oncol Lett. 2019 Sep;18(3):2240-2247. doi: 10.3892/ol.2019.10577. Epub 2019 Jul 5. Oncol Lett. 2019. PMID: 31404302 Free PMC article.
-
The oncogenic microRNA miR-222 promotes human LINE-1 retrotransposition.RNA Biol. 2025 Dec;22(1):1-15. doi: 10.1080/15476286.2025.2511318. Epub 2025 Jun 3. RNA Biol. 2025. PMID: 40421600 Free PMC article.
-
Antagonism between splicing and microprocessor complex dictates the serum-induced processing of lnc-MIRHG for efficient cell cycle reentry.RNA. 2020 Nov;26(11):1603-1620. doi: 10.1261/rna.075309.120. Epub 2020 Jul 16. RNA. 2020. PMID: 32675111 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

