Chronic Rhinosinusitis: Potential Role of Microbial Dysbiosis and Recommendations for Sampling Sites
- PMID: 29541629
- PMCID: PMC5836553
- DOI: 10.3389/fcimb.2018.00057
Chronic Rhinosinusitis: Potential Role of Microbial Dysbiosis and Recommendations for Sampling Sites
Abstract
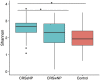
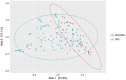
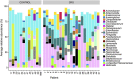
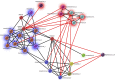
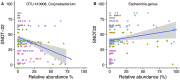
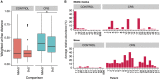
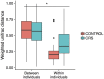
Chronic rhinosinusitis (CRS) is an inflammatory condition that affects up to 12% of the human population in developed countries. Previous studies examining the potential role of the sinus bacterial microbiota within CRS infections have found inconsistent results, possibly because of inconsistencies in sampling strategies. The aim of this study was to determine whether the sinus microbiome is altered in CRS and additionally if the middle meatus is a suitable representative site for sampling the sinus microbiome. Swab samples were collected from 12 healthy controls and 21 CRS patients, including all eight sinuses for CRS patients and between one and five sinuses for control subjects. The left and right middle meatus and nostril swabs were also collected. Significant differences in the sinus microbiomes between CRS and control samples were revealed using high-throughput 16S rRNA gene sequencing. The genus Escherichia was over-represented in CRS sinuses, and associations between control patients and Corynebacterium and Dolosigranulum were also identified. Comparisons of the middle meatuses between groups did not reflect these differences, and the abundance of the genus Escherichia was significantly lower at this location. Additionally, intra-patient variation was lower between sinuses than between sinus and middle meatus, which together with the above results suggests that the middle meatus is not an effective representative sampling site.
Keywords: 16S rRNA gene sequencing; chronic rhinosinusitis; microbiome; middle meatus; sinus.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

