A Novel Methodology for Characterizing Cell Subpopulations in Automated Time-lapse Microscopy
- PMID: 29541635
- PMCID: PMC5835524
- DOI: 10.3389/fbioe.2018.00017
A Novel Methodology for Characterizing Cell Subpopulations in Automated Time-lapse Microscopy
Abstract
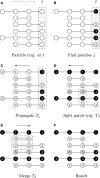
Time-lapse imaging of cell colonies in microfluidic chambers provides time series of bioimages, i.e., biomovies. They show the behavior of cells over time under controlled conditions. One of the main remaining bottlenecks in this area of research is the analysis of experimental data and the extraction of cell growth characteristics, such as lineage information. The extraction of the cell line by human observers is time-consuming and error-prone. Previously proposed methods often fail because of their reliance on the accurate detection of a single cell, which is not possible for high density, high diversity of cell shapes and numbers, and high-resolution images with high noise. Our task is to characterize subpopulations in biomovies. In order to shift the analysis of the data from individual cell level to cellular groups with similar fluorescence or even subpopulations, we propose to represent the cells by two new abstractions: the particle and the patch. We use a three-step framework: preprocessing, particle tracking, and construction of the patch lineage. First, preprocessing improves the signal-to-noise ratio and spatially aligns the biomovie frames. Second, cell sampling is performed by assuming particles, which represent a part of a cell, cell or group of contiguous cells in space. Particle analysis includes the following: particle tracking, trajectory linking, filtering, and color information, respectively. Particle tracking consists of following the spatiotemporal position of a particle and gives rise to coherent particle trajectories over time. Typical tracking problems may occur (e.g., appearance or disappearance of cells, spurious artifacts). They are effectively processed using trajectory linking and filtering. Third, the construction of the patch lineage consists in joining particle trajectories that share common attributes (i.e., proximity and fluorescence intensity) and feature common ancestry. This step is based on patch finding, patching trajectory propagation, patch splitting, and patch merging. The main idea is to group together the trajectories of particles in order to gain spatial coherence. The final result of CYCASP is the complete graph of the patch lineage. Finally, the graph encodes the temporal and spatial coherence of the development of cellular colonies. We present results showing a computation time of less than 5 min for biomovies and simulated films. The method, presented here, allowed for the separation of colonies into subpopulations and allowed us to interpret the growth of colonies in a timely manner.
Keywords: bacteria; bioimage informatics; bioimaging; cell lineage; image processing; microfluidics; synthetic biology.
Figures







Similar articles
-
Multi-feature-Based Robust Cell Tracking.Ann Biomed Eng. 2023 Mar;51(3):604-617. doi: 10.1007/s10439-022-03073-1. Epub 2022 Sep 14. Ann Biomed Eng. 2023. PMID: 36103061
-
Quantitative comparison of multiframe data association techniques for particle tracking in time-lapse fluorescence microscopy.Med Image Anal. 2015 Aug;24(1):163-189. doi: 10.1016/j.media.2015.06.006. Epub 2015 Jun 27. Med Image Anal. 2015. PMID: 26176413
-
Multiple dense particle tracking in fluorescence microscopy images based on multidimensional assignment.J Struct Biol. 2011 Feb;173(2):219-28. doi: 10.1016/j.jsb.2010.11.001. Epub 2010 Nov 10. J Struct Biol. 2011. PMID: 21073957
-
Analytics and visualization tools to characterize single-cell stochasticity using bacterial single-cell movie cytometry data.BMC Bioinformatics. 2021 Oct 29;22(1):531. doi: 10.1186/s12859-021-04409-9. BMC Bioinformatics. 2021. PMID: 34715773 Free PMC article.
-
Automatic three-dimensional segmentation of mouse embryonic stem cell nuclei by utilising multiple channels of confocal fluorescence images.J Microsc. 2021 Jan;281(1):57-75. doi: 10.1111/jmi.12949. Epub 2020 Aug 8. J Microsc. 2021. PMID: 32720710
Cited by
-
Perspectives on label-free microscopy of heterogeneous and dynamic biological systems.J Biomed Opt. 2025 Dec;29(Suppl 2):S22702. doi: 10.1117/1.JBO.29.S2.S22702. Epub 2024 Feb 29. J Biomed Opt. 2025. PMID: 38434231 Free PMC article.
-
SeeVis-3D space-time cube rendering for visualization of microfluidics image data.Bioinformatics. 2019 May 15;35(10):1802-1804. doi: 10.1093/bioinformatics/bty889. Bioinformatics. 2019. PMID: 30346487 Free PMC article.
-
Aardvark: Composite Visualizations of Trees, Time-Series, and Images.IEEE Trans Vis Comput Graph. 2025 Jan;31(1):1290-1300. doi: 10.1109/TVCG.2024.3456193. Epub 2024 Nov 25. IEEE Trans Vis Comput Graph. 2025. PMID: 39255114
-
Deciphering cell to cell spatial relationship for pathology images using SpatialQPFs.Sci Rep. 2024 Nov 28;14(1):29585. doi: 10.1038/s41598-024-81383-1. Sci Rep. 2024. PMID: 39609630 Free PMC article.
-
Computational Structural Biology: Successes, Future Directions, and Challenges.Molecules. 2019 Feb 12;24(3):637. doi: 10.3390/molecules24030637. Molecules. 2019. PMID: 30759724 Free PMC article. Review.
References
-
- Allan D., Caswell T., Keim N., van der Wel C. (2015). Trackpy: Trackpy v0.3.0. 10.5281/zenodo.34028 - DOI
-
- Bradski G. (2000). The OpenCV library. Dobb J. Softw. Tools. 120, 122–125.
-
- Crocker J., Grier D. (1996). Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179, 298–310.10.1006/jcis.1996.0217 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

