Rif1 phosphorylation site analysis in telomere length regulation and the response to damaged telomeres
- PMID: 29544213
- PMCID: PMC5911405
- DOI: 10.1016/j.dnarep.2018.03.001
Rif1 phosphorylation site analysis in telomere length regulation and the response to damaged telomeres
Abstract
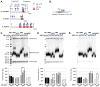
Telomeres, the ends of eukaryotic chromosomes, consist of repetitive DNA sequences and their bound proteins that protect the end from the DNA damage response. Short telomeres with fewer repeats are preferentially elongated by telomerase. Tel1, the yeast homolog of human ATM kinase, is preferentially recruited to short telomeres and Tel1 kinase activity is required for telomere elongation. Rif1, a telomere-binding protein, negatively regulates telomere length by forming a complex with two other telomere binding proteins, Rap1 and Rif2, to block telomerase recruitment. Rif1 has 14 SQ/TQ consensus phosphorylation sites for ATM kinases, including 6 in a SQ/TQ Cluster Domain (SCD) similar to other DNA damage response proteins. These 14 sites were analyzed as N-terminal, SCD and C-terminal domains. Mutating some sites to non-phosphorylatable residues increased telomere length in cells lacking Tel1 while a different set of phosphomimetic mutants increased telomere length in cells lacking Rif2, suggesting that Rif1 phosphorylation has both positive and negative effects on length regulation. While these mutations did not alter the sensitivity to DNA damaging agents, inducing telomere-specific damage by growing cells lacking YKU70 at high temperature revealed a role for the SCD. Mass spectrometry of Rif1 from wild type cells or those induced for telomere-specific DNA damage revealed increased phosphorylation in cells with telomere damage at an ATM consensus site in the SCD, S1351, and non-ATM sites S181 and S1637. A phosphomimetic rif1-S1351E mutation caused an increase in telomere length at synthetic telomeres but not natural telomeres. These results indicate that the Rif1 SCD can modulate Rif1 function. As all Rif1 orthologs have one or more SCD domains, these results for yeast Rif1 have implications for the regulation of Rif1 function in humans and other organisms.
Keywords: Phosphorylation; Rif1; Saccharomyces cerevisiae; Telomere length regulation.
Copyright © 2018 Elsevier B.V. All rights reserved.
Figures
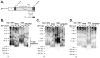



Similar articles
-
Rif1 and rif2 inhibit localization of tel1 to DNA ends.Mol Cell. 2009 Feb 13;33(3):312-22. doi: 10.1016/j.molcel.2008.12.027. Mol Cell. 2009. PMID: 19217405 Free PMC article.
-
At short telomeres Tel1 directs early replication and phosphorylates Rif1.PLoS Genet. 2014 Oct 16;10(10):e1004691. doi: 10.1371/journal.pgen.1004691. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25329891 Free PMC article.
-
Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae.Genetics. 1999 Aug;152(4):1531-41. doi: 10.1093/genetics/152.4.1531. Genetics. 1999. PMID: 10430581 Free PMC article.
-
Regulation of telomerase by telomeric proteins.Annu Rev Biochem. 2004;73:177-208. doi: 10.1146/annurev.biochem.73.071403.160049. Annu Rev Biochem. 2004. PMID: 15189140 Review.
-
Beyond interacting with Rap1: Dissecting the roles of Rif1.Int J Biol Macromol. 2025 May;306(Pt 3):141560. doi: 10.1016/j.ijbiomac.2025.141560. Epub 2025 Mar 1. Int J Biol Macromol. 2025. PMID: 40032092 Review.
Cited by
-
Rif1 regulates telomere length through conserved HEAT repeats.Nucleic Acids Res. 2021 Apr 19;49(7):3967-3980. doi: 10.1093/nar/gkab206. Nucleic Acids Res. 2021. PMID: 33772576 Free PMC article.
-
Proteomic analysis of bone marrow-derived mesenchymal stem cell extracellular vesicles from healthy donors: implications for proliferation, angiogenesis, Wnt signaling, and the basement membrane.Stem Cell Res Ther. 2021 Jun 5;12(1):328. doi: 10.1186/s13287-021-02405-7. Stem Cell Res Ther. 2021. PMID: 34090527 Free PMC article.
-
A CDK-Dependent Phosphorylation of a Novel Domain of Rif1 Regulates its Function during Telomere Damage and Other Types of Stress.Mol Cell Biol. 2023;43(5):185-199. doi: 10.1080/10985549.2023.2193768. Epub 2023 May 4. Mol Cell Biol. 2023. PMID: 37140180 Free PMC article.
-
RIF1 Links Replication Timing with Fork Reactivation and DNA Double-Strand Break Repair.Int J Mol Sci. 2021 Oct 23;22(21):11440. doi: 10.3390/ijms222111440. Int J Mol Sci. 2021. PMID: 34768871 Free PMC article. Review.
-
Shedding Light on the Interaction Between Rif1 and Telomeres in Ovarian Cancer.Aging Dis. 2024 Apr 1;15(2):535-545. doi: 10.14336/AD.2023.0716. Aging Dis. 2024. PMID: 37548940 Free PMC article. Review.
References
-
- Smogorzewska A, De Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. - PubMed
-
- Hardy CFJ, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. - PubMed
-
- Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

