Olaparib-induced Adaptive Response Is Disrupted by FOXM1 Targeting that Enhances Sensitivity to PARP Inhibition
- PMID: 29545475
- PMCID: PMC6705145
- DOI: 10.1158/1541-7786.MCR-17-0607
Olaparib-induced Adaptive Response Is Disrupted by FOXM1 Targeting that Enhances Sensitivity to PARP Inhibition
Abstract
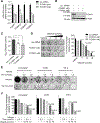
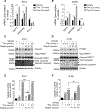
FOXM1 transcription factor network is activated in over 84% of cases in high-grade serous ovarian cancer (HGSOC), and FOXM1 upregulates the expression of genes involved in the homologous recombination (HR) DNA damage and repair (DDR) pathway. However, the role of FOXM1 in PARP inhibitor response has not yet been studied. This study demonstrates that PARP inhibitor (PARPi), olaparib, induces the expression and nuclear localization of FOXM1. On the basis of ChIP-qPCR, olaparib enhances the binding of FOXM1 to genes involved in HR repair. FOXM1 knockdown by RNAi or inhibition by thiostrepton decreases FOXM1 expression, decreases the expression of HR repair genes, such as BRCA1 and RAD51, and enhances sensitivity to olaparib. Comet and PARP trapping assays revealed increases in DNA damage and PARP trapping in FOXM1-inhibited cells treated with olaparib. Finally, thiostrepton decreases the expression of BRCA1 in rucaparib-resistant cells and enhances sensitivity to rucaparib. Collectively, these results identify that FOXM1 plays an important role in the adaptive response induced by olaparib and FOXM1 inhibition by thiostrepton induces "BRCAness" and enhances sensitivity to PARP inhibitors.Implications: FOXM1 inhibition represents an effective strategy to overcome resistance to PARPi, and targeting FOXM1-mediated adaptive pathways may produce better therapeutic effects for PARP inhibitors. Mol Cancer Res; 16(6); 961-73. ©2018 AACR.
©2018 American Association for Cancer Research.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures





Similar articles
-
BKM120 sensitizes BRCA-proficient triple negative breast cancer cells to olaparib through regulating FOXM1 and Exo1 expression.Sci Rep. 2021 Feb 26;11(1):4774. doi: 10.1038/s41598-021-82990-y. Sci Rep. 2021. PMID: 33637776 Free PMC article.
-
FDI-6 and olaparib synergistically inhibit the growth of pancreatic cancer by repressing BUB1, BRCA1 and CDC25A signaling pathways.Pharmacol Res. 2022 Jan;175:106040. doi: 10.1016/j.phrs.2021.106040. Epub 2021 Dec 22. Pharmacol Res. 2022. PMID: 34954029
-
Suberoylanilide hydroxamic acid (SAHA) enhances olaparib activity by targeting homologous recombination DNA repair in ovarian cancer.Gynecol Oncol. 2014 Jun;133(3):599-606. doi: 10.1016/j.ygyno.2014.03.007. Epub 2014 Mar 11. Gynecol Oncol. 2014. PMID: 24631446 Free PMC article.
-
Cediranib, a pan-VEGFR inhibitor, and olaparib, a PARP inhibitor, in combination therapy for high grade serous ovarian cancer.Expert Opin Investig Drugs. 2016;25(5):597-611. doi: 10.1517/13543784.2016.1156857. Epub 2016 Mar 16. Expert Opin Investig Drugs. 2016. PMID: 26899229 Review.
-
Clinical Application of Poly(ADP-Ribose) Polymerase Inhibitors in High-Grade Serous Ovarian Cancer.Oncologist. 2016 May;21(5):586-93. doi: 10.1634/theoncologist.2015-0438. Epub 2016 Mar 28. Oncologist. 2016. PMID: 27022037 Free PMC article. Review.
Cited by
-
The P72R Polymorphism in R248Q/W p53 Mutants Modifies the Mutant Effect on Epithelial to Mesenchymal Transition Phenotype and Cell Invasion via CXCL1 Expression.Int J Mol Sci. 2020 Oct 28;21(21):8025. doi: 10.3390/ijms21218025. Int J Mol Sci. 2020. PMID: 33126568 Free PMC article.
-
Clinical application of PARP inhibitors in ovarian cancer: from molecular mechanisms to the current status.J Ovarian Res. 2023 Jan 7;16(1):6. doi: 10.1186/s13048-023-01094-5. J Ovarian Res. 2023. PMID: 36611214 Free PMC article. Review.
-
The dynamic process of covalent and non-covalent PARylation in the maintenance of genome integrity: a focus on PARP inhibitors.NAR Cancer. 2023 Aug 21;5(3):zcad043. doi: 10.1093/narcan/zcad043. eCollection 2023 Sep. NAR Cancer. 2023. PMID: 37609662 Free PMC article.
-
FOXM1 Inhibition in Ovarian Cancer Tissue Cultures Affects Individual Treatment Susceptibility Ex Vivo.Cancers (Basel). 2021 Feb 25;13(5):956. doi: 10.3390/cancers13050956. Cancers (Basel). 2021. PMID: 33668819 Free PMC article.
-
Oncogenic B-Myb Is Associated With Deregulation of the DREAM-Mediated Cell Cycle Gene Expression Program in High Grade Serous Ovarian Carcinoma Clinical Tumor Samples.Front Oncol. 2021 Mar 4;11:637193. doi: 10.3389/fonc.2021.637193. eCollection 2021. Front Oncol. 2021. PMID: 33747961 Free PMC article.
References
-
- Giaccone G, Pinedo HM. Drug Resistance. Oncologist 1996;1:82–7. - PubMed
-
- Pfisterer J, Ledermann JA. Management of platinum-sensitive recurrent ovarian cancer. Semin Oncol 2006;33(2 Suppl 6):S12–S16. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

