Fusion of a highly N-glycosylated polypeptide increases the expression of ER-localized proteins in plants
- PMID: 29545574
- PMCID: PMC5854594
- DOI: 10.1038/s41598-018-22860-2
Fusion of a highly N-glycosylated polypeptide increases the expression of ER-localized proteins in plants
Abstract
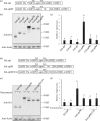
Plants represent promising systems for producing various recombinant proteins. One key area of focus for improving this technology is developing methods for producing recombinant proteins at high levels. Many methods have been developed to increase the transcript levels of recombinant genes. However, methods for increasing protein production involving steps downstream of transcription, including translation, have not been fully explored. Here, we investigated the effects of N-glycosylation on protein production and provide evidence that N-glycosylation greatly increases the expression levels of ER-targeted recombinant proteins. Fusion of the extracellular domain (M domain) of protein tyrosine phosphatase receptor type C (CD45), which contains four putative N-glycosylation sites to a model protein, leptin at the C-terminus, increased recombinant protein levels by 6.1 fold. This increase was specific to ER-targeted proteins and was dependent on N-glycosylation. Moreover, expression levels of leptin, leukemia inhibitory factor and GFP were also greatly increased by fusion of M domain at either the N or C-terminus. Furthermore, the increase in protein levels resulted from enhanced translation, but not transcription. Based on these results, we propose that fusing a small domain containing N-glycosylation sites to target proteins is a powerful technique for increasing the expression levels of recombinant proteins in plants.
Conflict of interest statement
The authors declare no competing interests.
Figures
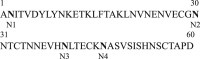

Similar articles
-
Endoplasmic reticulum retention motif fused to recombinant anti-cancer monoclonal antibody (mAb) CO17-1A affects mAb expression and plant stress response.PLoS One. 2018 Sep 24;13(9):e0198978. doi: 10.1371/journal.pone.0198978. eCollection 2018. PLoS One. 2018. PMID: 30248125 Free PMC article.
-
Protein body formation in stable transgenic tobacco expressing elastin-like polypeptide and hydrophobin fusion proteins.BMC Biotechnol. 2013 May 10;13:40. doi: 10.1186/1472-6750-13-40. BMC Biotechnol. 2013. PMID: 23663656 Free PMC article.
-
Light enhances the unfolded protein response as measured by BiP2 gene expression and the secretory GFP-2SC marker in Arabidopsis.Physiol Plant. 2008 Oct;134(2):360-8. doi: 10.1111/j.1399-3054.2008.01133.x. Epub 2008 Jun 28. Physiol Plant. 2008. PMID: 18494858
-
Biopharmaceuticals from plants: a multitude of options for posttranslational modifications.Biotechnol Genet Eng Rev. 2008;25:315-30. doi: 10.5661/bger-25-315. Biotechnol Genet Eng Rev. 2008. PMID: 21412360 Review.
-
Glyco-engineering of biotherapeutic proteins in plants.Mol Cells. 2008 Jun 30;25(4):494-503. Epub 2008 Apr 23. Mol Cells. 2008. PMID: 18443408 Review.
Cited by
-
A plant-produced SARS-CoV-2 spike protein elicits heterologous immunity in hamsters.Front Plant Sci. 2023 Mar 7;14:1146234. doi: 10.3389/fpls.2023.1146234. eCollection 2023. Front Plant Sci. 2023. PMID: 36959936 Free PMC article.
-
Production of active human iduronate-2-sulfatase (IDS) enzyme in Nicotiana benthamiana.Sci Rep. 2024 Oct 4;14(1):23066. doi: 10.1038/s41598-024-73778-x. Sci Rep. 2024. PMID: 39367006 Free PMC article.
-
Cost-effective production of tag-less recombinant protein in Nicotiana benthamiana.Plant Biotechnol J. 2019 Jun;17(6):1094-1105. doi: 10.1111/pbi.13040. Epub 2018 Dec 8. Plant Biotechnol J. 2019. PMID: 30468023 Free PMC article.
-
Production of Recombinant Active Human TGFβ1 in Nicotiana benthamiana.Front Plant Sci. 2022 May 31;13:922694. doi: 10.3389/fpls.2022.922694. eCollection 2022. Front Plant Sci. 2022. PMID: 35712604 Free PMC article.
-
Expression of kiwifruit-derived actinidin in Nicotiana benthamiana leaves.Front Plant Sci. 2025 Jan 10;15:1532170. doi: 10.3389/fpls.2024.1532170. eCollection 2024. Front Plant Sci. 2025. PMID: 39866318 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

