Secretome analysis of in vitro aged human mesenchymal stem cells reveals IGFBP7 as a putative factor for promoting osteogenesis
- PMID: 29545581
- PMCID: PMC5854613
- DOI: 10.1038/s41598-018-22855-z
Secretome analysis of in vitro aged human mesenchymal stem cells reveals IGFBP7 as a putative factor for promoting osteogenesis
Abstract
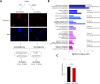
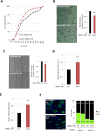
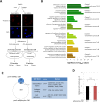
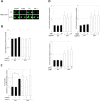
Aging is a complex biological process, which involves multiple mechanisms with different levels of regulation. Senescent cells are known to secrete senescence-associated proteins, which exert negative influences on surrounding cells. Mesenchymal stem cells (MSCs), the common progenitors for bone, cartilage and adipose tissue (which are especially affected tissues in aging), are known to secrete a broad spectrum of biologically active proteins with both paracrine and autocrine functions in many biological processes. In this report, we have studied the secreted factors (secretome) from human MSCs (hMSCs) and hMSCs-derived adipocytes which were induced to accumulate prelamin A, the immature form of the nuclear lamina protein called Lamin A, known to induce premature aging syndromes in humans and in murine models. Proteomic analysis from two different techniques, antibody arrays and LS-MS, showed that prelamin A accumulation in hMSCs promotes the differential secretion of factors previously identified as secreted by hMSCs undergoing osteogenesis. Moreover, this secretome was able to modulate osteogenesis of normal hMSCs in vitro. Finally, we found that one of the overexpressed secreted factors of this human aging in vitro stem cell model, IGFBP-7, is an osteogenic factor, essential for the viability of hMSCs during osteogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
GATA4-dependent regulation of the secretory phenotype via MCP-1 underlies lamin A-mediated human mesenchymal stem cell aging.Exp Mol Med. 2018 May 14;50(5):1-12. doi: 10.1038/s12276-018-0092-3. Exp Mol Med. 2018. PMID: 29760459 Free PMC article.
-
Rapid Induction of Osteogenic Markers in Mesenchymal Stem Cells by Adipose-Derived Stromal Vascular Fraction Cells.Cell Physiol Biochem. 2017;44(1):53-65. doi: 10.1159/000484582. Epub 2017 Nov 3. Cell Physiol Biochem. 2017. PMID: 29131029
-
IGFBP7 regulates the osteogenic differentiation of bone marrow-derived mesenchymal stem cells via Wnt/β-catenin signaling pathway.FASEB J. 2018 Apr;32(4):2280-2291. doi: 10.1096/fj.201700998RR. Epub 2018 Jan 5. FASEB J. 2018. PMID: 29242275
-
Crucial Role of Lamin A/C in the Migration and Differentiation of MSCs in Bone.Cells. 2020 May 26;9(6):1330. doi: 10.3390/cells9061330. Cells. 2020. PMID: 32466483 Free PMC article. Review.
-
Proteomic techniques for characterisation of mesenchymal stem cell secretome.Biochimie. 2013 Dec;95(12):2196-211. doi: 10.1016/j.biochi.2013.07.015. Epub 2013 Jul 20. Biochimie. 2013. PMID: 23880644 Review.
Cited by
-
IGFBP7 acts as a negative regulator of RANKL-induced osteoclastogenesis and oestrogen deficiency-induced bone loss.Cell Prolif. 2020 Feb;53(2):e12752. doi: 10.1111/cpr.12752. Epub 2019 Dec 30. Cell Prolif. 2020. PMID: 31889368 Free PMC article.
-
Serum-Free Manufacturing of Mesenchymal Stem Cell Tissue Rings Using Human-Induced Pluripotent Stem Cells.Stem Cells Int. 2019 Jan 15;2019:5654324. doi: 10.1155/2019/5654324. eCollection 2019. Stem Cells Int. 2019. PMID: 30766604 Free PMC article.
-
Paracrine senescence of human endometrial mesenchymal stem cells: a role for the insulin-like growth factor binding protein 3.Aging (Albany NY). 2020 Jan 17;12(2):1987-2004. doi: 10.18632/aging.102737. Epub 2020 Jan 17. Aging (Albany NY). 2020. PMID: 31951594 Free PMC article.
-
Transcriptomic Profiling of Adipose Derived Stem Cells Undergoing Osteogenesis by RNA-Seq.Sci Rep. 2019 Aug 13;9(1):11800. doi: 10.1038/s41598-019-48089-1. Sci Rep. 2019. PMID: 31409848 Free PMC article.
-
Reprogramming of human fibroblasts into osteoblasts by insulin-like growth factor-binding protein 7.Stem Cells Transl Med. 2020 Mar;9(3):403-415. doi: 10.1002/sctm.19-0281. Epub 2020 Jan 6. Stem Cells Transl Med. 2020. PMID: 31904196 Free PMC article.
References
-
- Ruiz de Eguino G, et al. Sp1 transcription factor interaction with accumulated prelamin a impairs adipose lineage differentiation in human mesenchymal stem cells: essential role of sp1 in the integrity of lipid vesicles. Stem Cells Transl Med. 2012;1:309–321. doi: 10.5966/sctm.2011-0010. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

