Antiviral Effect of Ribavirin against HCV Associated with Increased Frequency of G-to-A and C-to-U Transitions in Infectious Cell Culture Model
- PMID: 29545599
- PMCID: PMC5854589
- DOI: 10.1038/s41598-018-22620-2
Antiviral Effect of Ribavirin against HCV Associated with Increased Frequency of G-to-A and C-to-U Transitions in Infectious Cell Culture Model
Abstract
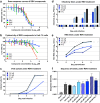
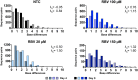
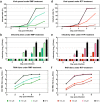
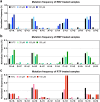
Ribavirin (RBV) is a broad-spectrum antiviral active against a wide range of RNA viruses. Despite having been used for decades in the treatment of chronic hepatitis C virus (HCV) infection, the precise mechanism of action of RBV is unknown. In other viruses, it inhibits propagation by increasing the rate of G-to-A and C-to-U transitions. Here, we utilized the J6/JFH1 HCV cell-culture system to investigate whether RBV inhibits HCV through the same mechanism. Infected Huh7.5 cells were treated with increasing concentrations of RBV or its phosphorylated forms. A fragment of the HCV NS5B-polymerase gene was amplified, cloned, and sequenced to estimate genetic distances. We confirm that the antiviral effect of all three RBV-drug forms on HCV relies on induction of specific transitions (G-to-A and C-to-U). These mutations lead to generation of non-infectious virions, reflected by decreased spread of HCV in cell culture despite relatively limited effect on virus genome titers. Moreover, treatment experiments conducted on a novel Huh7.5 cell line stably overexpressing adenosine kinase, a key enzyme for RBV activation, yielded comparable results. This study indicates that RBV action on HCV in hepatoma cell-culture is exerted through increase in mutagenesis, mediated by RBV triphosphate, and leading to production of non-infectious viruses.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- AASLD-IDSA. HCV Guidance: Recommendations for Testing, Managing, and Treating Hepatitis C, http://www.hcvguidelines.org (2018). - PubMed
-
- EASL Recommendations on Treatment of Hepatitis C 2016. J Hepatol66, 153–194, 10.1016/j.jhep.2016.09.001 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

