Repertoire Analysis of Antibody CDR-H3 Loops Suggests Affinity Maturation Does Not Typically Result in Rigidification
- PMID: 29545810
- PMCID: PMC5840193
- DOI: 10.3389/fimmu.2018.00413
Repertoire Analysis of Antibody CDR-H3 Loops Suggests Affinity Maturation Does Not Typically Result in Rigidification
Abstract
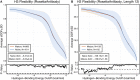
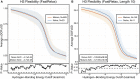
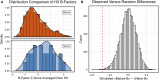
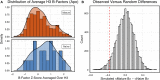
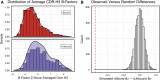
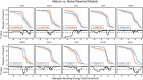
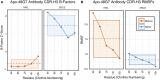
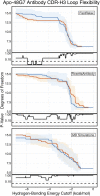
Antibodies can rapidly evolve in specific response to antigens. Affinity maturation drives this evolution through cycles of mutation and selection leading to enhanced antibody specificity and affinity. Elucidating the biophysical mechanisms that underlie affinity maturation is fundamental to understanding B-cell immunity. An emergent hypothesis is that affinity maturation reduces the conformational flexibility of the antibody's antigen-binding paratope to minimize entropic losses incurred upon binding. In recent years, computational and experimental approaches have tested this hypothesis on a small number of antibodies, often observing a decrease in the flexibility of the complementarity determining region (CDR) loops that typically comprise the paratope and in particular the CDR-H3 loop, which contributes a plurality of antigen contacts. However, there were a few exceptions and previous studies were limited to a small handful of cases. Here, we determined the structural flexibility of the CDR-H3 loop for thousands of recent homology models of the human peripheral blood cell antibody repertoire using rigidity theory. We found no clear delineation in the flexibility of naïve and antigen-experienced antibodies. To account for possible sources of error, we additionally analyzed hundreds of human and mouse antibodies in the Protein Data Bank through both rigidity theory and B-factor analysis. By both metrics, we observed only a slight decrease in the CDR-H3 loop flexibility when comparing affinity matured antibodies to naïve antibodies, and the decrease was not as drastic as previously reported. Further analysis, incorporating molecular dynamics simulations, revealed a spectrum of changes in flexibility. Our results suggest that rigidification may be just one of many biophysical mechanisms for increasing affinity.
Keywords: RosettaAntibody; affinity maturation; antibody repertoires; complementarity determining regions; conformational flexibility; molecular dynamics simulations; pebble game algorithm; rigidity theory.
Figures
Similar articles
-
Multi-constraint computational design suggests that native sequences of germline antibody H3 loops are nearly optimal for conformational flexibility.Proteins. 2009 Jun;75(4):846-58. doi: 10.1002/prot.22293. Proteins. 2009. PMID: 19194863 Free PMC article.
-
CDR-H3 diversity is not required for antigen recognition by synthetic antibodies.J Mol Biol. 2013 Feb 22;425(4):803-11. doi: 10.1016/j.jmb.2012.11.037. Epub 2012 Dec 3. J Mol Biol. 2013. PMID: 23219464 Free PMC article.
-
Predicting antibody complementarity determining region structures without classification.Mol Biosyst. 2011 Dec;7(12):3327-34. doi: 10.1039/c1mb05223c. Epub 2011 Oct 20. Mol Biosyst. 2011. PMID: 22011953
-
The sequences encoded by immunoglobulin diversity (DH ) gene segments play key roles in controlling B-cell development, antigen-binding site diversity, and antibody production.Immunol Rev. 2018 Jul;284(1):106-119. doi: 10.1111/imr.12669. Immunol Rev. 2018. PMID: 29944758 Review.
-
Diversity in the Cow Ultralong CDR H3 Antibody Repertoire.Front Immunol. 2018 Jun 4;9:1262. doi: 10.3389/fimmu.2018.01262. eCollection 2018. Front Immunol. 2018. PMID: 29915599 Free PMC article. Review.
Cited by
-
Design of nanobody-based bispecific constructs by in silico affinity maturation and umbrella sampling simulations.Comput Struct Biotechnol J. 2022 Dec 16;21:601-613. doi: 10.1016/j.csbj.2022.12.021. eCollection 2023. Comput Struct Biotechnol J. 2022. PMID: 36659922 Free PMC article.
-
Structural modeling of antibody variable regions using deep learning-progress and perspectives on drug discovery.Front Mol Biosci. 2023 Jul 7;10:1214424. doi: 10.3389/fmolb.2023.1214424. eCollection 2023. Front Mol Biosci. 2023. PMID: 37484529 Free PMC article. Review.
-
Multistate design of influenza antibodies improves affinity and breadth against seasonal viruses.Proc Natl Acad Sci U S A. 2019 Jan 29;116(5):1597-1602. doi: 10.1073/pnas.1806004116. Epub 2019 Jan 14. Proc Natl Acad Sci U S A. 2019. PMID: 30642961 Free PMC article.
-
Computational design of structured loops for new protein functions.Biol Chem. 2019 Feb 25;400(3):275-288. doi: 10.1515/hsz-2018-0348. Biol Chem. 2019. PMID: 30676995 Free PMC article. Review.
-
Breaking the law: unconventional strategies for antibody diversification.Nat Rev Immunol. 2019 Jun;19(6):355-368. doi: 10.1038/s41577-019-0126-7. Nat Rev Immunol. 2019. PMID: 30718829 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

