DNA methylation is a common molecular alteration in colorectal cancer cells and culture method has no influence on DNA methylation
- PMID: 29545832
- PMCID: PMC5841015
- DOI: 10.3892/etm.2018.5809
DNA methylation is a common molecular alteration in colorectal cancer cells and culture method has no influence on DNA methylation
Abstract
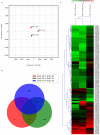
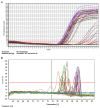
The present study aimed to explore whether culture method had an influence on DNA methylation in colorectal cancer (CRC). In the present study, CRC cells were cultured in two-dimensional (2D), three-dimensional (3D) and mouse orthotopic transplantation (Tis) cultures. Principal component analysis (PCA) was used for global visualization of the three samples. A Venn diagram was applied for intersection and union analysis for different comparisons. The methylation condition of 5'-C-phosphate-G-3' (CpG) location was determined using unsupervised clustering analysis. Scatter plots and histograms of the mean β values between 3D vs. 2D, 3D vs. Tis and Tis vs. 2D were constructed. In order to explore the biological function of the genes, gene ontology and Kyoto Encyclopedia of Gene and Genomes (KEGG) pathway analyses were utilized. To explore the influence of culture condition on genes, quantitative methylation specific polymerase chain reaction (QMSP) was performed. The three samples connected with each other closely, as demonstrated by PCA. Venn diagram analysis indicated that some differential methylation positions were commonly shared in the three groups of samples and 16 CpG positions appeared hypermethylated in the three samples. The methylation patterns between the 3D and 2D cultures were more similar than those of 3D and Tis, and Tis and 2D. Results of gene ontology demonstrated that differentially expressed genes were involved in molecular function, cellular components and biological function. KEGG analysis indicated that genes were enriched in 13 pathways, of which four pathways were the most evident. These pathways were pathways in cancer, mitogen-activated protein kinase signaling, axon guidance and insulin signaling. Furthermore, QMSP demonstrated that methylation of mutL homolog, phosphatase and tensin homolog, runt-related transcription factor, Ras association family member, cadherin-1, O-6-methylguanine-DNA-methyltransferase and P16 genes had no obvious difference in 2D, 3D and Tis culture conditions. In conclusion, the culture method had no influence on DNA methylation in CRC cells.
Keywords: 5′-C-phosphate-G-3′; DNA methylation; colorectal cancer; culture method.
Figures


Similar articles
-
DNA methylation is related to the occurrence of breast cancer and is not affected by culture conditions.Mol Med Rep. 2018 May;17(5):7365-7371. doi: 10.3892/mmr.2018.8735. Epub 2018 Mar 14. Mol Med Rep. 2018. PMID: 29568926
-
Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis.World J Gastroenterol. 2004 Dec 1;10(23):3441-54. doi: 10.3748/wjg.v10.i23.3441. World J Gastroenterol. 2004. PMID: 15526363 Free PMC article.
-
Gene methylation in gastric cancer.Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
-
Alteration in the expression of MGMT and RUNX3 due to non-CpG promoter methylation and their correlation with different risk factors in esophageal cancer patients.Tumour Biol. 2017 May;39(5):1010428317701630. doi: 10.1177/1010428317701630. Tumour Biol. 2017. PMID: 28468586
-
Epigenetic regulation in human melanoma: past and future.Epigenetics. 2015;10(2):103-21. doi: 10.1080/15592294.2014.1003746. Epigenetics. 2015. PMID: 25587943 Free PMC article. Review.
Cited by
-
Methylenetetrahydrofolate Dehydrogenase 1 Silencing Expedites the Apoptosis of Non-Small Cell Lung Cancer Cells via Modulating DNA Methylation.Med Sci Monit. 2018 Oct 21;24:7499-7507. doi: 10.12659/MSM.910265. Med Sci Monit. 2018. PMID: 30343310 Free PMC article.
-
Comparative analysis between 2D and 3D colorectal cancer culture models for insights into cellular morphological and transcriptomic variations.Sci Rep. 2023 Oct 26;13(1):18380. doi: 10.1038/s41598-023-45144-w. Sci Rep. 2023. PMID: 37884554 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
