Comprehensive analysis of differential expression profiles reveals potential biomarkers associated with the cell cycle and regulated by p53 in human small cell lung cancer
- PMID: 29545845
- PMCID: PMC5841087
- DOI: 10.3892/etm.2018.5833
Comprehensive analysis of differential expression profiles reveals potential biomarkers associated with the cell cycle and regulated by p53 in human small cell lung cancer
Abstract
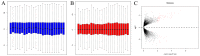
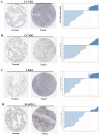
Small cell lung cancer (SCLC) is the subtype of lung cancer with the highest degree of malignancy and the lowest degree of differentiation. The purpose of this study was to investigate the molecular mechanisms of SCLC using bioinformatics analysis, and to provide new ideas for the early diagnosis and targeted therapy of SCLC. Microarray data were downloaded from Gene Expression Omnibus. Differentially expressed genes (DEGs) in SCLC were compared with the normal lung samples and identified. Gene Ontology (GO) function and pathway analysis of DEGs was performed through the DAVID database. Furthermore, microarray data was analyzed by using the clustering analysis tool GoMiner. Protein-protein interaction (PPI) networks of DEGs were constructed using the STRING online database. Protein expression was determined from the Human Protein Atlas, and SCLC gene expression was determined using Oncomine. In total, 153 DEGs were obtained. Functional enrichment analysis suggested that the majority of DEGs were associated with the cell cycle. CCNB1, CCNB2, MAD2L1 and CDK1 were identified to contribute to the progression of SCLC through combined use of GO, Kyoto Encyclopedia of Genes and Genomes enrichment analysis and a PPI network. mRNA and protein expression were also validated in an integrative database. The present study indicated that the formation of SCLC may be associated with cell cycle regulation. In addition, the four crucial genes CCNB1, CCNB2, MAD2L1 and CDK1, which are downstream of p53, may have important roles in the occurrence and progression of SCLC, and thus may be promising potential biomarkers and therapeutic targets.
Keywords: DAVID; cell cycle; function enrichment; microarray analysis; small cell lung cancer.
Figures




Similar articles
-
Identification of candidate genes associated with the pathogenesis of small cell lung cancer via integrated bioinformatics analysis.Oncol Lett. 2019 Oct;18(4):3723-3733. doi: 10.3892/ol.2019.10685. Epub 2019 Jul 29. Oncol Lett. 2019. PMID: 31516585 Free PMC article.
-
Identification of candidate biomarkers and pathways associated with SCLC by bioinformatics analysis.Mol Med Rep. 2018 Aug;18(2):1538-1550. doi: 10.3892/mmr.2018.9095. Epub 2018 May 29. Mol Med Rep. 2018. PMID: 29845250 Free PMC article.
-
Identification and Integrated Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer.Med Sci Monit. 2019 Dec 5;25:9280-9289. doi: 10.12659/MSM.918620. Med Sci Monit. 2019. PMID: 31805030 Free PMC article.
-
Identification of potential therapeutic targets for colorectal cancer by bioinformatics analysis.Oncol Lett. 2016 Dec;12(6):5092-5098. doi: 10.3892/ol.2016.5328. Epub 2016 Oct 31. Oncol Lett. 2016. PMID: 28105216 Free PMC article.
-
Screening Hub Genes as Prognostic Biomarkers of Hepatocellular Carcinoma by Bioinformatics Analysis.Cell Transplant. 2019 Dec;28(1_suppl):76S-86S. doi: 10.1177/0963689719893950. Epub 2019 Dec 11. Cell Transplant. 2019. PMID: 31822116 Free PMC article.
Cited by
-
Identification of candidate genes associated with the pathogenesis of small cell lung cancer via integrated bioinformatics analysis.Oncol Lett. 2019 Oct;18(4):3723-3733. doi: 10.3892/ol.2019.10685. Epub 2019 Jul 29. Oncol Lett. 2019. PMID: 31516585 Free PMC article.
-
Identification of potential agents for thymoma by integrated analyses of differentially expressed tumour-associated genes and molecular docking experiments.Exp Ther Med. 2019 Sep;18(3):2001-2014. doi: 10.3892/etm.2019.7817. Epub 2019 Jul 26. Exp Ther Med. 2019. PMID: 31452699 Free PMC article.
-
Lung Cancer: One Disease or Many.Hum Hered. 2018;83(2):65-70. doi: 10.1159/000488942. Epub 2018 Jun 5. Hum Hered. 2018. PMID: 29864749 Free PMC article.
-
Identification of potential target genes and crucial pathways in small cell lung cancer based on bioinformatic strategy and human samples.PLoS One. 2020 Nov 13;15(11):e0242194. doi: 10.1371/journal.pone.0242194. eCollection 2020. PLoS One. 2020. PMID: 33186389 Free PMC article.
-
Identifying General Tumor and Specific Lung Cancer Biomarkers by Transcriptomic Analysis.Biology (Basel). 2022 Jul 20;11(7):1082. doi: 10.3390/biology11071082. Biology (Basel). 2022. PMID: 36101460 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
