Investigating a multigene prognostic assay based on significant pathways for Luminal A breast cancer through gene expression profile analysis
- PMID: 29545900
- PMCID: PMC5840762
- DOI: 10.3892/ol.2018.7940
Investigating a multigene prognostic assay based on significant pathways for Luminal A breast cancer through gene expression profile analysis
Abstract
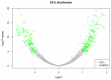
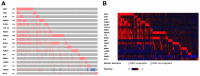
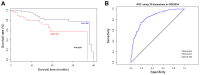
The present study aimed to investigate potential recurrence-risk biomarkers based on significant pathways for Luminal A breast cancer through gene expression profile analysis. Initially, the gene expression profiles of Luminal A breast cancer patients were downloaded from The Cancer Genome Atlas database. The differentially expressed genes (DEGs) were identified using a Limma package and the hierarchical clustering analysis was conducted for the DEGs. In addition, the functional pathways were screened using Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses and rank ratio calculation. The multigene prognostic assay was exploited based on the statistically significant pathways and its prognostic function was tested using train set and verified using the gene expression data and survival data of Luminal A breast cancer patients downloaded from the Gene Expression Omnibus. A total of 300 DEGs were identified between good and poor outcome groups, including 176 upregulated genes and 124 downregulated genes. The DEGs may be used to effectively distinguish Luminal A samples with different prognoses verified by hierarchical clustering analysis. There were 9 pathways screened as significant pathways and a total of 18 DEGs involved in these 9 pathways were identified as prognostic biomarkers. According to the survival analysis and receiver operating characteristic curve, the obtained 18-gene prognostic assay exhibited good prognostic function with high sensitivity and specificity to both the train and test samples. In conclusion the 18-gene prognostic assay including the key genes, transcription factor 7-like 2, anterior parietal cortex and lymphocyte enhancer factor-1 may provide a new method for predicting outcomes and may be conducive to the promotion of precision medicine for Luminal A breast cancer.
Keywords: Luminal A breast cancer; differentially expressed genes; multigene prognostic assay; significant pathways.
Figures


Similar articles
-
Bioinformatics analyses of significant genes, related pathways and candidate prognostic biomarkers in glioblastoma.Mol Med Rep. 2018 Nov;18(5):4185-4196. doi: 10.3892/mmr.2018.9411. Epub 2018 Aug 21. Mol Med Rep. 2018. PMID: 30132538 Free PMC article.
-
Bioinformatics analysis of gene expression profiles to identify causal genes in luminal B2 breast cancer.Oncol Lett. 2017 Dec;14(6):7880-7888. doi: 10.3892/ol.2017.7256. Epub 2017 Oct 23. Oncol Lett. 2017. PMID: 29250180 Free PMC article.
-
Bioinformatics analysis of gene expression profile data to screen key genes involved in intracranial aneurysms.Mol Med Rep. 2019 Nov;20(5):4415-4424. doi: 10.3892/mmr.2019.10696. Epub 2019 Sep 23. Mol Med Rep. 2019. PMID: 31545495 Free PMC article.
-
Identification of Methylation Markers and Differentially Expressed Genes with Prognostic Value in Breast Cancer.J Comput Biol. 2019 Dec;26(12):1394-1408. doi: 10.1089/cmb.2019.0179. Epub 2019 Jul 10. J Comput Biol. 2019. PMID: 31290690
-
Prognostic biomarkers related to breast cancer recurrence identified based on Logit model analysis.World J Surg Oncol. 2020 Sep 25;18(1):254. doi: 10.1186/s12957-020-02026-z. World J Surg Oncol. 2020. PMID: 32977823 Free PMC article.
Cited by
-
The role of C1orf50 in breast cancer progression and prognosis.Breast Cancer. 2025 Mar;32(2):292-305. doi: 10.1007/s12282-024-01653-8. Epub 2024 Nov 28. Breast Cancer. 2025. PMID: 39604563 Free PMC article.
References
-
- Aure MR, Vitelli V, Jernström S, Kumar S, Krohn M, Due EU, Haukaas TH, Leivonen SK, Vollan HK, Lüders T, et al. Integrative clustering reveals a novel split in the Luminal A subtype of breast cancer with impact on outcome. Breast Cancer Res. 2017;19:44. doi: 10.1186/s13058-017-0812-y. - DOI - PMC - PubMed
-
- Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, Senn HJ., Panel members Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. - DOI - PMC - PubMed
-
- Jagsi R, Raad RA, Goldberg S, Sullivan T, Michaelson J, Powell SN, Taghian AG. Locoregional recurrence rates and prognostic factors for failure in node-negative patients treated with mastectomy: Implications for postmastectomy radiation. Int J Radiat Oncol Biol Phys. 2005;62:1035–1039. doi: 10.1016/j.ijrobp.2004.12.014. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
