Divergence in problem-solving skills is associated with differential expression of glutamate receptors in wild finches
- PMID: 29546239
- PMCID: PMC5851658
- DOI: 10.1126/sciadv.aao6369
Divergence in problem-solving skills is associated with differential expression of glutamate receptors in wild finches
Abstract
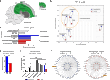
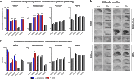
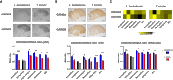
Problem solving and innovation are key components of intelligence. We compare wild-caught individuals from two species that are close relatives of Darwin's finches, the innovative Loxigilla barbadensis, and its most closely related species in Barbados, the conservative Tiaris bicolor. We found an all-or-none difference in the problem-solving capacity of the two species. Brain RNA sequencing analyses revealed interspecific differences in genes related to neuronal and synaptic plasticity in the intrapallial neural populations (mesopallium and nidopallium), especially in the nidopallium caudolaterale, a structure functionally analogous to the mammalian prefrontal cortex. At a finer scale, we discovered robust differences in glutamate receptor expression between the species. In particular, the GRIN2B/GRIN2A ratio, known to correlate with synaptic plasticity, was higher in the innovative L. barbadensis. These findings suggest that divergence in avian intelligence is associated with similar neuronal mechanisms to that of mammals, including humans.
Figures

References
-
- Timmermans S., Lefebvre L., Boire D., Basu P., Relative size of the hyperstriatum ventrale is the best predictor of feeding innovation rate in birds. Brain Behav. Evol. 56, 196–203 (2000). - PubMed
-
- Mehlhorn J., Hunt G. R., Gray R. D., Rehkämper G., Güntürkün O., Tool-making New Caledonian crows have large associative brain areas. Brain Behav. Evol. 75, 63–70 (2010). - PubMed
-
- Mace G. M., Harvey P. H., Clutton-Brock T. H., Is brain size an ecological variable? Trends Neurosci. 3, 193–196 (1980).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

