A complete logical approach to resolve the evolution and dynamics of mitochondrial genome in bilaterians
- PMID: 29547666
- PMCID: PMC5856267
- DOI: 10.1371/journal.pone.0194334
A complete logical approach to resolve the evolution and dynamics of mitochondrial genome in bilaterians
Abstract
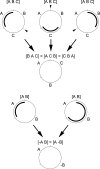
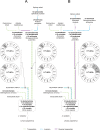
Investigating how recombination might modify gene order during the evolution has become a routine part of mitochondrial genome analysis. A new method of genomic maps analysis based on formal logic is described. The purpose of this method is to 1) use mitochondrial gene order of current taxa as datasets 2) calculate rearrangements between all mitochondrial gene orders and 3) reconstruct phylogenetic relationships according to these calculated rearrangements within a tree under the assumption of maximum parsimony. Unlike existing methods mainly based on the probabilistic approach, the main strength of this new approach is that it calculates all the exact tree solutions with completeness and provides logical consequences as highly robust results. Moreover, this method infers all possible hypothetical ancestors and reconstructs character states for all internal nodes of the trees. We started by testing our method using the deuterostomes as a study case. Then, with sponges as an outgroup, we investigated the evolutionary history of mitochondrial genomes of 47 bilaterian phyla and emphasised the peculiar case of chaetognaths. This pilot work showed that the use of formal logic in a hypothetico-deductive background such as phylogeny (where experimental testing of hypotheses is impossible) is very promising to explore mitochondrial gene order in deuterostomes and should be applied to many other bilaterian clades.
Conflict of interest statement
Figures


Similar articles
-
A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data.Mol Phylogenet Evol. 2008 Oct;49(1):59-68. doi: 10.1016/j.ympev.2008.07.004. Epub 2008 Jul 15. Mol Phylogenet Evol. 2008. PMID: 18672078
-
Complete mitochondrial genomes reveal phylogeny relationship and evolutionary history of the family Felidae.Genet Mol Res. 2013 Sep 3;12(3):3256-62. doi: 10.4238/2013.September.3.1. Genet Mol Res. 2013. PMID: 24065666
-
The complete mitochondrial genome of the onychophoran Epiperipatus biolleyi reveals a unique transfer RNA set and provides further support for the ecdysozoa hypothesis.Mol Biol Evol. 2008 Jan;25(1):42-51. doi: 10.1093/molbev/msm223. Epub 2007 Oct 13. Mol Biol Evol. 2008. PMID: 17934206
-
Bioinformatics methods for the comparative analysis of metazoan mitochondrial genome sequences.Mol Phylogenet Evol. 2013 Nov;69(2):320-7. doi: 10.1016/j.ympev.2012.09.019. Epub 2012 Sep 28. Mol Phylogenet Evol. 2013. PMID: 23023207 Review.
-
Genome trees and the nature of genome evolution.Annu Rev Microbiol. 2005;59:191-209. doi: 10.1146/annurev.micro.59.030804.121233. Annu Rev Microbiol. 2005. PMID: 16153168 Review.
Cited by
-
Enhanced dynamicity: evolutionary insights into amphibian mitogenomes architecture.BMC Genomics. 2025 Mar 17;26(1):261. doi: 10.1186/s12864-025-11480-6. BMC Genomics. 2025. PMID: 40097969 Free PMC article.
-
qGO: a novel method for quantifying the diversity of mitochondrial genome organization.BMC Genomics. 2024 Nov 18;25(1):1097. doi: 10.1186/s12864-024-11006-6. BMC Genomics. 2024. PMID: 39558268 Free PMC article.
-
Mitogenomes Reveal Alternative Initiation Codons and Lineage-Specific Gene Order Conservation in Echinoderms.Mol Biol Evol. 2021 Mar 9;38(3):981-985. doi: 10.1093/molbev/msaa262. Mol Biol Evol. 2021. PMID: 33027524 Free PMC article.
-
Architectural instability, inverted skews and mitochondrial phylogenomics of Isopoda: outgroup choice affects the long-branch attraction artefacts.R Soc Open Sci. 2020 Feb 5;7(2):191887. doi: 10.1098/rsos.191887. eCollection 2020 Feb. R Soc Open Sci. 2020. PMID: 32257344 Free PMC article.
References
-
- Moritz C, Dowling TE, Brown WM. Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annual Rev Ecol Syst. 1987; 18:269–292.
-
- Boore JL, Brown WM. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev. 1998; 8(6):668–674. - PubMed
-
- Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999; 33:351–397. doi: 10.1146/annurev.genet.33.1.351 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

