A novel transcript isoform of STING that sequesters cGAMP and dominantly inhibits innate nucleic acid sensing
- PMID: 29547894
- PMCID: PMC5934658
- DOI: 10.1093/nar/gky186
A novel transcript isoform of STING that sequesters cGAMP and dominantly inhibits innate nucleic acid sensing
Abstract
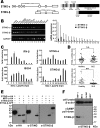
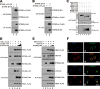
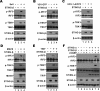
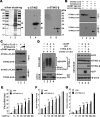
STING is a core adaptor in innate nucleic acid sensing in mammalian cells, on which different sensing pathways converge to induce type I interferon (IFN) production. Particularly, STING is activated by 2'3'-cGAMP, a cyclic dinucleotide containing mixed phosphodiester linkages and produced by cytoplasmic DNA sensor cGAS. Here, we reported on a novel transcript isoform of STING designated STING-β that dominantly inhibits innate nucleic acid sensing. STING-β without transmembrane domains was widely expressed at low levels in various human tissues and viral induction of STING-β correlated inversely with IFN-β production. The expression of STING-β declined in patients with lupus, in which type I IFNs are commonly overproduced. STING-β suppressed the induction of IFNs, IFN-stimulated genes and other cytokines by various immunostimulatory agents including cyclic dinucleotides, DNA, RNA and viruses, whereas depletion of STING-β showed the opposite effect. STING-β interacted with STING-α and antagonized its antiviral function. STING-β also interacted with TBK1 and prevented it from binding with STING-α, TRIF or other transducers. In addition, STING-β bound to 2'3'-cGAMP and impeded its binding with and activation of STING-α, leading to suppression of IFN-β production. Taken together, STING-β sequesters 2'3'-cGAMP second messenger and other transducer molecules to inhibit innate nucleic acid sensing dominantly.
Figures



Similar articles
-
African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the cGAS-STING Pathway.J Virol. 2019 May 29;93(12):e02298-18. doi: 10.1128/JVI.02298-18. Print 2019 Jun 15. J Virol. 2019. PMID: 30918080 Free PMC article.
-
TBK1 and IKKε Act Redundantly to Mediate STING-Induced NF-κB Responses in Myeloid Cells.Cell Rep. 2020 Apr 7;31(1):107492. doi: 10.1016/j.celrep.2020.03.056. Cell Rep. 2020. PMID: 32268090
-
The cGas-Sting Signaling Pathway Is Required for the Innate Immune Response Against Ectromelia Virus.Front Immunol. 2018 Jun 14;9:1297. doi: 10.3389/fimmu.2018.01297. eCollection 2018. Front Immunol. 2018. PMID: 29963044 Free PMC article.
-
cGAS-STING Pathway's Impact on Intestinal Barrier.J Gastroenterol Hepatol. 2025 Jun;40(6):1381-1392. doi: 10.1111/jgh.16974. Epub 2025 May 16. J Gastroenterol Hepatol. 2025. PMID: 40377214 Review.
-
When STING Meets Viruses: Sensing, Trafficking and Response.Front Immunol. 2020 Sep 29;11:2064. doi: 10.3389/fimmu.2020.02064. eCollection 2020. Front Immunol. 2020. PMID: 33133062 Free PMC article. Review.
Cited by
-
Protocol for the identification and expression analysis of a cytoplasmic membrane-localized protein STING.STAR Protoc. 2023 Mar 20;4(2):102172. doi: 10.1016/j.xpro.2023.102172. Online ahead of print. STAR Protoc. 2023. PMID: 36943863 Free PMC article.
-
Genome-Wide Analysis of Alternative Splicing during Host-Virus Interactions in Chicken.Viruses. 2021 Dec 2;13(12):2409. doi: 10.3390/v13122409. Viruses. 2021. PMID: 34960678 Free PMC article.
-
Inflammatory responses revealed through HIV infection of microglia-containing cerebral organoids.J Neuroinflammation. 2025 Feb 10;22(1):36. doi: 10.1186/s12974-025-03353-2. J Neuroinflammation. 2025. PMID: 39930449 Free PMC article.
-
Self-assembled cGAMP-STINGΔTM signaling complex as a bioinspired platform for cGAMP delivery.Sci Adv. 2020 Jun 12;6(24):eaba7589. doi: 10.1126/sciadv.aba7589. eCollection 2020 Jun. Sci Adv. 2020. PMID: 32582856 Free PMC article.
-
African Swine Fever Virus EP364R and C129R Target Cyclic GMP-AMP To Inhibit the cGAS-STING Signaling Pathway.J Virol. 2022 Aug 10;96(15):e0102222. doi: 10.1128/jvi.01022-22. Epub 2022 Jul 21. J Virol. 2022. PMID: 35861515 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

