Defect-facilitated buckling in supercoiled double-helix DNA
- PMID: 29548184
- PMCID: PMC5940003
- DOI: 10.1103/PhysRevE.97.022416
Defect-facilitated buckling in supercoiled double-helix DNA
Abstract
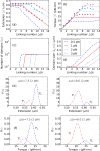
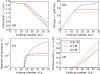
We present a statistical-mechanical model for stretched twisted double-helix DNA, where thermal fluctuations are treated explicitly from a Hamiltonian without using any scaling hypotheses. Our model applied to defect-free supercoiled DNA describes the coexistence of multiple plectoneme domains in long DNA molecules at physiological salt concentrations (≈0.1M Na^{+}) and stretching forces (≈1pN). We find a higher (lower) number of domains at lower (higher) ionic strengths and stretching forces, in accord with experimental observations. We use our model to study the effect of an immobile point defect on the DNA contour that allows a localized kink. The degree of the kink is controlled by the defect size, such that a larger defect further reduces the bending energy of the defect-facilitated kinked end loop. We find that a defect can spatially pin a plectoneme domain via nucleation of a kinked end loop, in accord with experiments and simulations. Our model explains previously reported magnetic tweezer experiments [A. Dittmore et al., Phys. Rev. Lett. 119, 147801 (2017)PRLTAO0031-900710.1103/PhysRevLett.119.147801] showing two buckling signatures: buckling and "rebuckling" in supercoiled DNA with a base-unpaired region. Comparing with experiments, we find that under 1 pN force, a kinked end loop nucleated at a base-mismatched site reduces the bending energy by ≈0.7 k_{B}T per unpaired base. Our model predicts the coexistence of three states at the buckling and rebuckling transitions, which warrants new experiments.
Figures





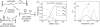





Similar articles
-
Torque and buckling in stretched intertwined double-helix DNAs.Phys Rev E. 2017 May;95(5-1):052401. doi: 10.1103/PhysRevE.95.052401. Epub 2017 May 1. Phys Rev E. 2017. PMID: 28618488 Free PMC article.
-
Energetics at the DNA supercoiling transition.Biophys J. 2010 Apr 7;98(7):1267-76. doi: 10.1016/j.bpj.2009.12.4292. Biophys J. 2010. PMID: 20371326 Free PMC article.
-
Nucleation of Multiple Buckled Structures in Intertwined DNA Double Helices.Phys Rev Lett. 2017 Nov 3;119(18):188103. doi: 10.1103/PhysRevLett.119.188103. Epub 2017 Oct 31. Phys Rev Lett. 2017. PMID: 29219598 Free PMC article.
-
Conformational and thermodynamic properties of supercoiled DNA.Annu Rev Biophys Biomol Struct. 1994;23:609-43. doi: 10.1146/annurev.bb.23.060194.003141. Annu Rev Biophys Biomol Struct. 1994. PMID: 7919794 Review.
-
Through the looking glass: the discovery of supercoiled DNA.Trends Biochem Sci. 1990 May;15(5):202-7. doi: 10.1016/0968-0004(90)90163-6. Trends Biochem Sci. 1990. PMID: 2193440 Review.
Cited by
-
DNA supercoiling-mediated collective behavior of co-transcribing RNA polymerases.Nucleic Acids Res. 2022 Feb 22;50(3):1269-1279. doi: 10.1093/nar/gkab1252. Nucleic Acids Res. 2022. PMID: 34951454 Free PMC article.
-
Physical principles of retroviral integration in the human genome.Nat Commun. 2019 Feb 4;10(1):575. doi: 10.1038/s41467-019-08333-8. Nat Commun. 2019. PMID: 30718508 Free PMC article.
-
Investigating site-selection mechanisms of retroviral integration in supercoiled DNA braids.J R Soc Interface. 2021 Aug;18(181):20210229. doi: 10.1098/rsif.2021.0229. Epub 2021 Aug 25. J R Soc Interface. 2021. PMID: 34428944 Free PMC article.
-
DNA fluctuations reveal the size and dynamics of topological domains.PNAS Nexus. 2022 Nov 22;1(5):pgac268. doi: 10.1093/pnasnexus/pgac268. eCollection 2022 Nov. PNAS Nexus. 2022. PMID: 36712371 Free PMC article.
-
Coarse-grained modelling of DNA plectoneme pinning in the presence of base-pair mismatches.Nucleic Acids Res. 2020 Nov 4;48(19):10713-10725. doi: 10.1093/nar/gkaa836. Nucleic Acids Res. 2020. PMID: 33045724 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
