OsNOA1 functions in a threshold-dependent manner to regulate chloroplast proteins in rice at lower temperatures
- PMID: 29548275
- PMCID: PMC5857130
- DOI: 10.1186/s12870-018-1258-9
OsNOA1 functions in a threshold-dependent manner to regulate chloroplast proteins in rice at lower temperatures
Abstract
Background: Although decreased protein expressions have been observed in NOA1 (Nitric Oxide Associated protein 1) deficient plants, the molecular mechanisms of how NOA1 regulates protein metabolism remain poorly understood. In this study, we have used a global comparative proteomic approach for both OsNOA1 suppression and overexpression transgenic lines under two different temperatures, in combination with physiological and biochemical analyses to explore the regulatory mechanisms of OsNOA1 in rice.
Results: In OsNOA1-silenced or highly overexpressed rice, considerably different expression patterns of both chlorophyll and Rubisco as well as distinct phenotypes were observed between the growth temperatures at 22 °C and 30 °C. These observations led us to hypothesize there appears a narrow abundance threshold for OsNOA1 to function properly at lower temperatures, while higher temperatures seem to partially compensate for the changes of OsNOA1 abundance. Quantitative proteomic analyses revealed higher temperatures could restore 90% of the suppressed proteins to normal levels, whereas almost all of the remaining suppressed proteins were chloroplast ribosomal proteins. Additionally, our data showed 90% of the suppressed proteins in both types of transgenic plants at lower temperatures were located in the chloroplast, suggesting a primary effect of OsNOA1 on chloroplast proteins. Transcript analyses, along with in vitro pull-down experiments further demonstrated OsNOA1 is associated with the function of chloroplast ribosomes.
Conclusions: Our results suggest OsNOA1 functions in a threshold-dependent manner for regulation of chloroplast proteins at lower temperatures, which may be mediated by interactions between OsNOA1 and chloroplast ribosomes.
Keywords: Chloroplast; NOA1; Quantitative proteomics; Rice; Tandem mass tag; Threshold-dependent.
Conflict of interest statement
Ethics approval and consent to participate
This study does not contain any research requiring ethical consent or approval.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



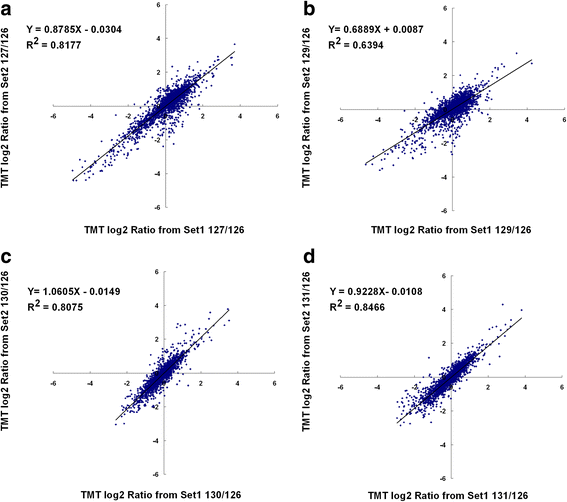


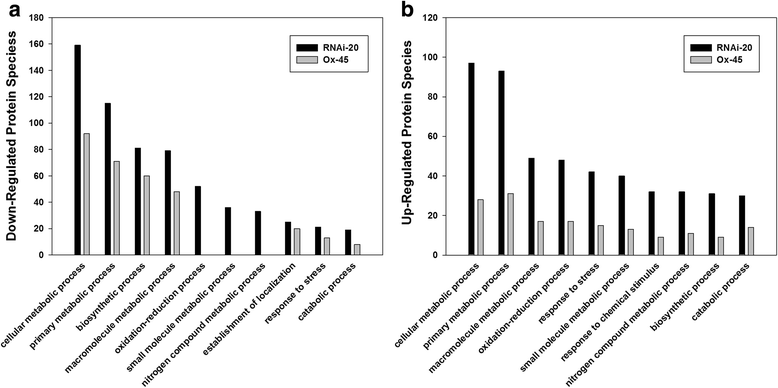


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

