Total RNA extraction from tissues for microRNA and target gene expression analysis: not all kits are created equal
- PMID: 29548320
- PMCID: PMC5857145
- DOI: 10.1186/s12896-018-0421-6
Total RNA extraction from tissues for microRNA and target gene expression analysis: not all kits are created equal
Abstract
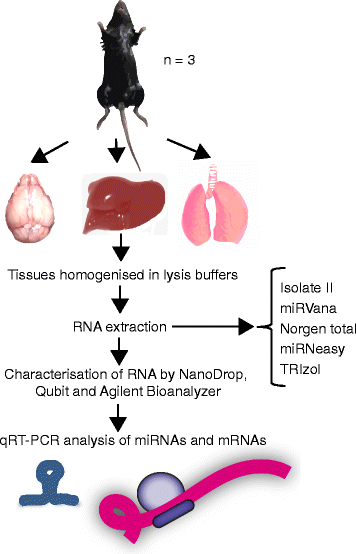
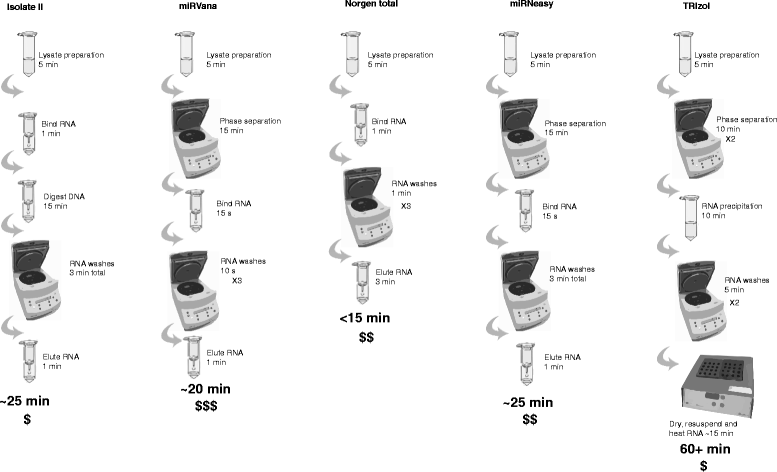
Background: microRNAs (miRNAs) are short non-coding RNAs that fine-tune gene expression. The aberrant expression of miRNAs is associated with many diseases and they have both therapeutic and biomarker potential. However, our understanding of their usefulness is dependent on the tools we have to study them. Previous studies have identified the need to optimise and standardise RNA extraction methods in order to avoid biased results. Herein, we extracted RNA from murine lung, liver and brain tissues using five commercially available total RNA extraction methods. These included either: phenol: chloroform extraction followed by alcohol precipitation (TRIzol), phenol:chloroform followed by solid-phase extraction (column-based; miRVana and miRNeasy) and solid-phase separation with/without affinity resin (Norgen total and Isolate II). We then evaluated each extraction method for the quality and quantity of RNA recovered, and the expression of miRNAs and target genes.
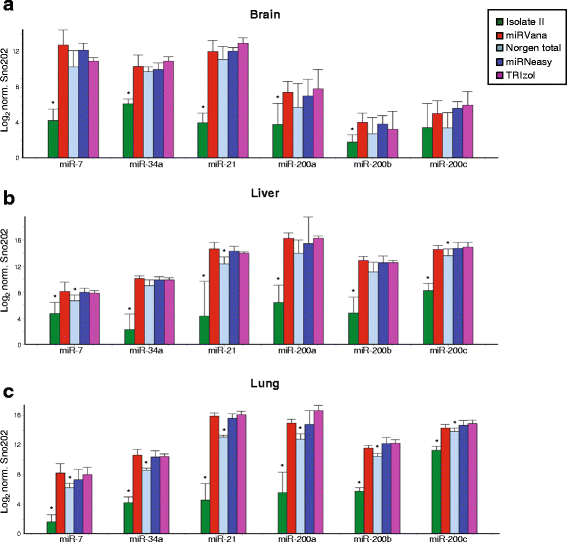
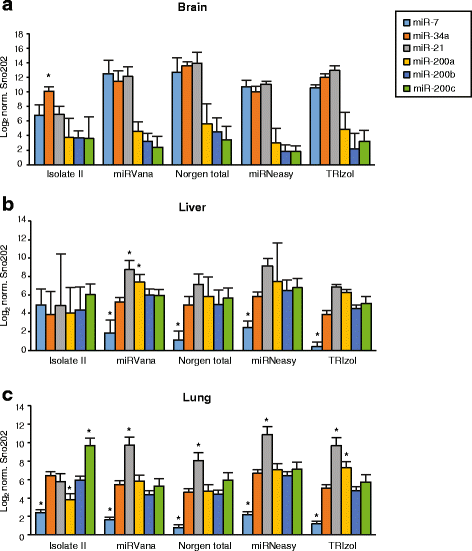
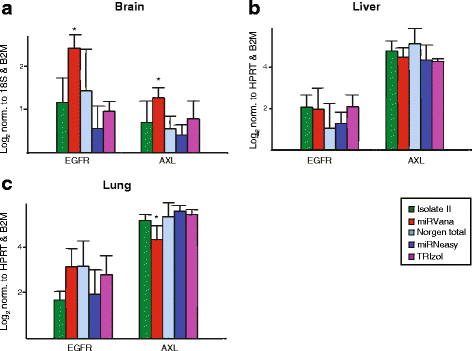
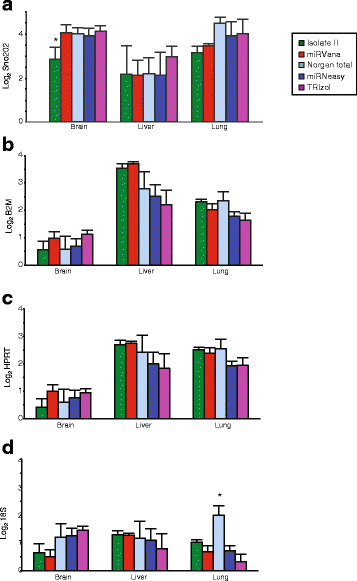
Results: We identified differences between each of the RNA extraction methods in the quantity and quality of RNA samples, and in the analysis of miRNA and target gene expression. For the purposes of consistency in quantity, quality and high recovery of miRNAs from tissues, we identified that Phenol:chloroform phase separation combined with silica column-based solid extraction method was preferable (miRVana microRNA isolation). We also identified a method that is not appropriate for miRNA analysis from tissue samples (Bioline Isolate II). For target gene expression any of the kits could be used to analyse mRNA, but if interested in analysing mRNA and miRNA from the same RNA samples some methods should be avoided.
Conclusions: Different methods used to isolate miRNAs will yield different results and therefore a robust RNA isolation method is required for reproducibility. Researchers should optimise these methods for their specific application and keep in mind that "total RNA" extraction methods do not isolate all types of RNA equally.
Keywords: Biomarkers; Extraction; Real-time PCR; Tissue; miRNA-based therapy; microRNA isolation method; microRNAs.
Conflict of interest statement
Ethics approval
Animals were sourced, housed and treated in accordance with institutional guidelines approved by the Harry Perkins Institute of Medical Research Ethics Committee under application #AEO35.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

