Haplotype-Based Genome-Wide Prediction Models Exploit Local Epistatic Interactions Among Markers
- PMID: 29549092
- PMCID: PMC5940160
- DOI: 10.1534/g3.117.300548
Haplotype-Based Genome-Wide Prediction Models Exploit Local Epistatic Interactions Among Markers
Abstract
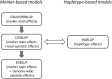
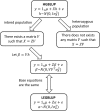
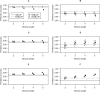
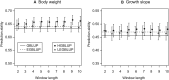
Genome-wide prediction approaches represent versatile tools for the analysis and prediction of complex traits. Mostly they rely on marker-based information, but scenarios have been reported in which models capitalizing on closely-linked markers that were combined into haplotypes outperformed marker-based models. Detailed comparisons were undertaken to reveal under which circumstances haplotype-based genome-wide prediction models are superior to marker-based models. Specifically, it was of interest to analyze whether and how haplotype-based models may take local epistatic effects between markers into account. Assuming that populations consisted of fully homozygous individuals, a marker-based model in which local epistatic effects inside haplotype blocks were exploited (LEGBLUP) was linearly transformable into a haplotype-based model (HGBLUP). This theoretical derivation formally revealed that haplotype-based genome-wide prediction models capitalize on local epistatic effects among markers. Simulation studies corroborated this finding. Due to its computational efficiency the HGBLUP model promises to be an interesting tool for studies in which ultra-high-density SNP data sets are studied. Applying the HGBLUP model to empirical data sets revealed higher prediction accuracies than for marker-based models for both traits studied using a mouse panel. In contrast, only a small subset of the traits analyzed in crop populations showed such a benefit. Cases in which higher prediction accuracies are observed for HGBLUP than for marker-based models are expected to be of immediate relevance for breeders, due to the tight linkage a beneficial haplotype will be preserved for many generations. In this respect the inheritance of local epistatic effects very much resembles the one of additive effects.
Keywords: GenPred; Genomic Selection; Shared Data Resources; epistasis; genome-wide prediction; haplotype; local epistatic effect.
Copyright © 2018 Jiang et al.
Figures


References
-
- Boichard D., Guillaume F., Baur A., Croiseau P., Rossignol M. N., et al. , 2012. Genomic selection in French dairy cattle. Anim. Prod. Sci. 52(3): 115–120. 10.1071/AN11119 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
