Calcineurin-mediated IL-2 production by CD11chighMHCII+ myeloid cells is crucial for intestinal immune homeostasis
- PMID: 29549257
- PMCID: PMC5856784
- DOI: 10.1038/s41467-018-03495-3
Calcineurin-mediated IL-2 production by CD11chighMHCII+ myeloid cells is crucial for intestinal immune homeostasis
Abstract
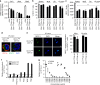
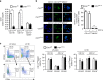
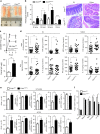
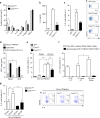
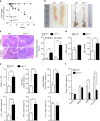
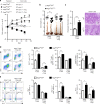
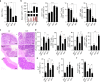
The intestinal immune system can respond to invading pathogens yet maintain immune tolerance to self-antigens and microbiota. Myeloid cells are central to these processes, but the signaling pathways that underlie tolerance versus inflammation are unclear. Here we show that mice lacking Calcineurin B in CD11chighMHCII+ cells (Cnb1 CD11c mice) spontaneously develop intestinal inflammation and are susceptible to induced colitis. In these mice, colitis is associated with expansion of T helper type 1 (Th1) and Th17 cell populations and a decrease in the number of FoxP3+ regulatory T (Treg) cells, and the pathology is linked to the inability of intestinal Cnb1-deficient CD11chighMHCII+ cells to express IL-2. Deleting IL-2 in CD11chighMHCII+ cells induces spontaneous colitis resembling human inflammatory bowel disease. Our findings identify that the calcineurin-NFAT-IL-2 pathway in myeloid cells is a critical regulator of intestinal homeostasis by influencing the balance of inflammatory and regulatory responses in the mouse intestine.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Calcineurin B in CD4+ T Cells Prevents Autoimmune Colitis by Negatively Regulating the JAK/STAT Pathway.Front Immunol. 2018 Feb 19;9:261. doi: 10.3389/fimmu.2018.00261. eCollection 2018. Front Immunol. 2018. PMID: 29515579 Free PMC article.
-
IL-10 control of CD11c+ myeloid cells is essential to maintain immune homeostasis in the small and large intestine.Oncotarget. 2016 May 31;7(22):32015-30. doi: 10.18632/oncotarget.8337. Oncotarget. 2016. PMID: 27027442 Free PMC article.
-
Interferon-γ induces expression of MHC class II on intestinal epithelial cells and protects mice from colitis.PLoS One. 2014 Jan 28;9(1):e86844. doi: 10.1371/journal.pone.0086844. eCollection 2014. PLoS One. 2014. PMID: 24489792 Free PMC article.
-
[Regulation of the human gut homeostasis by anti-inflammatory CD14+ CD163high CD160high myeloid cells].Nihon Rinsho Meneki Gakkai Kaishi. 2016;39(5):441-447. doi: 10.2177/jsci.39.441. Nihon Rinsho Meneki Gakkai Kaishi. 2016. PMID: 27795500 Review. Japanese.
-
The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin.Annu Rev Pathol. 2013 Jan 24;8:477-512. doi: 10.1146/annurev-pathol-011110-130318. Epub 2012 Nov 15. Annu Rev Pathol. 2013. PMID: 23157335 Free PMC article. Review.
Cited by
-
CD301b+ dendritic cell-derived IL-2 dictates CD4+ T helper cell differentiation.Nat Commun. 2025 Feb 26;16(1):2002. doi: 10.1038/s41467-025-55916-9. Nat Commun. 2025. PMID: 40011469 Free PMC article.
-
Duan-Nai-An, A Yeast Probiotic, Improves Intestinal Mucosa Integrity and Immune Function in Weaned Piglets.Sci Rep. 2020 Mar 12;10(1):4556. doi: 10.1038/s41598-020-61279-6. Sci Rep. 2020. PMID: 32165666 Free PMC article.
-
USP16-mediated deubiquitination of calcineurin A controls peripheral T cell maintenance.J Clin Invest. 2019 May 28;129(7):2856-2871. doi: 10.1172/JCI123801. eCollection 2019 May 28. J Clin Invest. 2019. PMID: 31135381 Free PMC article.
-
Inositol 1,4,5-trisphosphate 3-kinase B promotes Ca2+ mobilization and the inflammatory activity of dendritic cells.Sci Signal. 2021 Mar 30;14(676):eaaz2120. doi: 10.1126/scisignal.aaz2120. Sci Signal. 2021. PMID: 33785611 Free PMC article.
-
Calcineurin inhibitors reduce NFAT-dependent expression of antifungal pentraxin-3 by human monocytes.J Leukoc Biol. 2020 Mar;107(3):497-508. doi: 10.1002/JLB.4VMA0318-138R. Epub 2019 Apr 1. J Leukoc Biol. 2020. PMID: 30934147 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

