Tat controls transcriptional persistence of unintegrated HIV genome in primary human macrophages
- PMID: 29549786
- PMCID: PMC6021179
- DOI: 10.1016/j.virol.2018.03.006
Tat controls transcriptional persistence of unintegrated HIV genome in primary human macrophages
Abstract
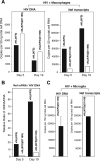
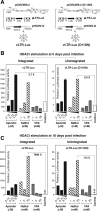
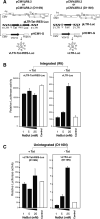
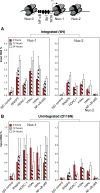
In HIV infected macrophages, a large population of viral genomes persists as the unintegrated form (uDNA) that is transcriptionally active. However, how this transcriptional activity is controlled remains unclear. In this report, we investigated whether Tat, the viral transactivator of transcription, is involved in uDNA transcription. We demonstrate that de novo Tat activity is generated from uDNA, and this uDNA-derived Tat (uTat) transactivates the uDNA LTR. In addition, uTat is required for the transcriptional persistence of uDNA that is assembled into repressive episomal minichromatin. In the absence of uTat, uDNA minichromatin is gradually silenced, but remains highly inducible by HDAC inhibitors (HDACi). Therefore, functionally, uTat antagonizes uDNA minichromatin repression to maintain persistent viral transcription in macrophages. uTat-mediated viral persistence may establish a viral reservoir in macrophages where uDNA were found to persist.
Keywords: HDACi; HIV; Macrophage; Persistence; Reservoir; Tat; Transcription; Unintegrated HIV DNA.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Aguilar-Cordova E, Chinen J, Donehower L, Lewis DE, Belmont JW. A sensitive reporter cell line for HIV-1 tat activity, HIV-1 inhibitors, and T cell activation effects. AIDS Res Hum Retroviruses. 1994;10:295–301. - PubMed
-
- Akan E, Chang-Liu CM, Watanabe J, Ishizawa K, Woloschak GE. The effects of vinblastine on the expression of human immunodeficiency virus type 1 long terminal repeat. Leuk Res. 1997;21:459–464. - PubMed
-
- Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, Nakatani Y, Jeang KT. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem. 1998;273:24898–24905. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

