Hosts and Sources of Endemic Human Coronaviruses
- PMID: 29551135
- PMCID: PMC7112090
- DOI: 10.1016/bs.aivir.2018.01.001
Hosts and Sources of Endemic Human Coronaviruses
Abstract
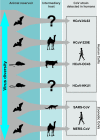
The four endemic human coronaviruses HCoV-229E, -NL63, -OC43, and -HKU1 contribute a considerable share of upper and lower respiratory tract infections in adults and children. While their clinical representation resembles that of many other agents of the common cold, their evolutionary histories, and host associations could provide important insights into the natural history of past human pandemics. For two of these viruses, we have strong evidence suggesting an origin in major livestock species while primordial associations for all four viruses may have existed with bats and rodents. HCoV-NL63 and -229E may originate from bat reservoirs as assumed for many other coronaviruses, but HCoV-OC43 and -HKU1 seem more likely to have speciated from rodent-associated viruses. HCoV-OC43 is thought to have emerged from ancestors in domestic animals such as cattle or swine. The bovine coronavirus has been suggested to be a possible ancestor, from which HCoV-OC43 may have emerged in the context of a pandemic recorded historically at the end of the 19th century. New data suggest that HCoV-229E may actually be transferred from dromedary camels similar to Middle East respiratory syndrome (MERS) coronavirus. This scenario provides important ecological parallels to the present prepandemic pattern of host associations of the MERS coronavirus.
Keywords: Alphacoronavirus; Betacoronavirus; Chiroptera; Coronaviridae; Livestock; Respiratory tract infections; Rodentia; Zoonotic diseases.
© 2018 Elsevier Inc. All rights reserved.
Figures


References
-
- Alekseev K.P., Vlasova A.N., Jung K., Hasoksuz M., Zhang X., Halpin R., Wang S., Ghedin E., Spiro D., Saif L.J. Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences. J. Virol. 2008;82:12422–12431. - PMC - PubMed
-
- Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y.A., Kalko E.K.V., Lina P.H.C., Godlevska E.V., Reusken C., Seebens A., Gloza-Rausch F., Vallo P., Tschapka M., Drosten C., Drexler J.F. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19:456–459. - PMC - PubMed
-
- Annan A., Ebach F., Corman V.M., Krumkamp R., Adu-Sarkodie Y., Eis-Hubinger A.M., Kruppa T., Simon A., May J., Evans J., Panning M., Drosten C., Drexler J.F. Similar virus spectra and seasonality in paediatric patients with acute respiratory disease, Ghana and Germany. Clin. Microbiol. Infect. 2016;22:340–346. - PMC - PubMed
-
- Anthony S.J., Gilardi K., Menachery V.D., Goldstein T., Ssebide B., Mbabazi R., Navarrete-Macias I., Liang E., Wells H., Hicks A., Petrosov A., Byarugaba D.K., Debbink K., Dinnon K.H., Scobey T., Randell S.H., Yount B.L., Cranfield M., Johnson C.K., Baric R.S., Lipkin W.I., Mazet J.A. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. mBio. 2017;8(2) e00373-17. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

