Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing
- PMID: 29551268
- PMCID: PMC5866243
- DOI: 10.1016/j.cell.2018.02.034
Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing
Abstract
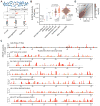
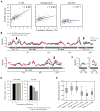
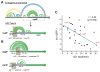
mRNAs can fold into complex structures that regulate gene expression. Resolving such structures de novo has remained challenging and has limited our understanding of the prevalence and functions of mRNA structure. We use SHAPE-MaP experiments in living E. coli cells to derive quantitative, nucleotide-resolution structure models for 194 endogenous transcripts encompassing approximately 400 genes. Individual mRNAs have exceptionally diverse architectures, and most contain well-defined structures. Active translation destabilizes mRNA structure in cells. Nevertheless, mRNA structure remains similar between in-cell and cell-free environments, indicating broad potential for structure-mediated gene regulation. We find that the translation efficiency of endogenous genes is regulated by unfolding kinetics of structures overlapping the ribosome binding site. We discover conserved structured elements in 35% of UTRs, several of which we validate as novel protein binding motifs. RNA structure regulates every gene studied here in a meaningful way, implying that most functional structures remain to be discovered.
Keywords: RNA binding proteins; RNA structure; non-coding RNA; ribosomal proteins; translation efficiency; translation regulation; translational coupling.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
K.M.W. is an advisor to and holds equity in Ribometrix, to which SHAPE-MaP technologies have been licensed.
Figures




Comment in
-
Structure in sequence.Nat Chem Biol. 2018 May;14(5):413. doi: 10.1038/s41589-018-0050-3. Nat Chem Biol. 2018. PMID: 29662185 No abstract available.
Similar articles
-
Structured mRNAs regulate translation initiation by binding to the platform of the ribosome.Cell. 2007 Sep 21;130(6):1019-31. doi: 10.1016/j.cell.2007.07.008. Cell. 2007. PMID: 17889647
-
Trapping the ribosome to control gene expression.Cell. 2007 Sep 21;130(6):983-5. doi: 10.1016/j.cell.2007.09.002. Cell. 2007. PMID: 17889642
-
Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA.J Mol Biol. 2003 Aug 22;331(4):737-43. doi: 10.1016/s0022-2836(03)00809-x. J Mol Biol. 2003. PMID: 12909006
-
Optimizing scaleup yield for protein production: Computationally Optimized DNA Assembly (CODA) and Translation Engineering.Biotechnol Annu Rev. 2007;13:27-42. doi: 10.1016/S1387-2656(07)13002-7. Biotechnol Annu Rev. 2007. PMID: 17875472 Review.
-
[A model for trans-translation].Yi Chuan. 2006 Aug;28(8):1051-4. Yi Chuan. 2006. PMID: 16870596 Review. Chinese.
Cited by
-
Ribosome recycling is not critical for translational coupling in Escherichia coli.Elife. 2020 Sep 23;9:e59974. doi: 10.7554/eLife.59974. Elife. 2020. PMID: 32965213 Free PMC article.
-
From reporters to endogenous genes: the impact of the first five codons on translation efficiency in Escherichia coli.RNA Biol. 2019 Dec;16(12):1806-1816. doi: 10.1080/15476286.2019.1661213. Epub 2019 Sep 5. RNA Biol. 2019. PMID: 31470761 Free PMC article.
-
RNA base-pairing complexity in living cells visualized by correlated chemical probing.Proc Natl Acad Sci U S A. 2019 Dec 3;116(49):24574-24582. doi: 10.1073/pnas.1905491116. Epub 2019 Nov 19. Proc Natl Acad Sci U S A. 2019. PMID: 31744869 Free PMC article.
-
Probing RNA structure in vivo.Curr Opin Struct Biol. 2019 Dec;59:151-158. doi: 10.1016/j.sbi.2019.07.008. Epub 2019 Sep 13. Curr Opin Struct Biol. 2019. PMID: 31521910 Free PMC article. Review.
-
Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing.Elife. 2020 Feb 17;9:e55002. doi: 10.7554/eLife.55002. Elife. 2020. PMID: 32065583 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

