A Dehydration-Induced Eukaryotic Translation Initiation Factor iso4G Identified in a Slow Wilting Soybean Cultivar Enhances Abiotic Stress Tolerance in Arabidopsis
- PMID: 29552022
- PMCID: PMC5840855
- DOI: 10.3389/fpls.2018.00262
A Dehydration-Induced Eukaryotic Translation Initiation Factor iso4G Identified in a Slow Wilting Soybean Cultivar Enhances Abiotic Stress Tolerance in Arabidopsis
Abstract
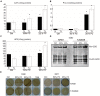
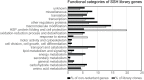
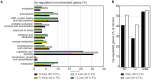
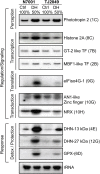
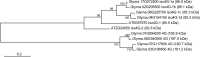
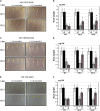
Water is usually the main limiting factor for soybean productivity worldwide and yet advances in genetic improvement for drought resistance in this crop are still limited. In the present study, we investigated the physiological and molecular responses to drought in two soybean contrasting genotypes, a slow wilting N7001 and a drought sensitive TJS2049 cultivars. Measurements of stomatal conductance, carbon isotope ratios and accumulated dry matter showed that N7001 responds to drought by employing mechanisms resulting in a more efficient water use than TJS2049. To provide an insight into the molecular mechanisms that these cultivars employ to deal with water stress, their early and late transcriptional responses to drought were analyzed by suppression subtractive hybridization. A number of differentially regulated genes from N7001 were identified and their expression pattern was compared between in this genotype and TJS2049. Overall, the data set indicated that N7001 responds to drought earlier than TJ2049 by up-regulating a larger number of genes, most of them encoding proteins with regulatory and signaling functions. The data supports the idea that at least some of the phenotypic differences between slow wilting and drought sensitive plants may rely on the regulation of the level and timing of expression of specific genes. One of the genes that exhibited a marked N7001-specific drought induction profile encoded a eukaryotic translation initiation factor iso4G (GmeIFiso4G-1a). GmeIFiso4G-1a is one of four members of this protein family in soybean, all of them sharing high sequence identity with each other. In silico analysis of GmeIFiso4G-1 promoter sequences suggested a possible functional specialization between distinct family members, which can attain differences at the transcriptional level. Conditional overexpression of GmeIFiso4G-1a in Arabidopsis conferred the transgenic plants increased tolerance to osmotic, salt, drought and low temperature stress, providing a strong experimental evidence for a direct association between a protein of this class and general abiotic stress tolerance mechanisms. Moreover, the results of this work reinforce the importance of the control of protein synthesis as a central mechanism of stress adaptation and opens up for new strategies for improving crop performance under stress.
Keywords: Arabidopsis; abiotic stress; drought; eIFiso4G; soybean; translation initiation.
Figures


Similar articles
-
Functional specialization of chloroplast vesiculation (CV) duplicated genes from soybean shows partial overlapping roles during stress-induced or natural senescence.Front Plant Sci. 2023 Jun 6;14:1184020. doi: 10.3389/fpls.2023.1184020. eCollection 2023. Front Plant Sci. 2023. PMID: 37346131 Free PMC article.
-
Genome-wide transcriptional analysis of two soybean genotypes under dehydration and rehydration conditions.BMC Genomics. 2013 Oct 6;14:687. doi: 10.1186/1471-2164-14-687. BMC Genomics. 2013. PMID: 24093224 Free PMC article.
-
The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants.Plant Mol Biol. 2011 Apr;75(6):537-53. doi: 10.1007/s11103-011-9738-4. Epub 2011 Feb 18. Plant Mol Biol. 2011. PMID: 21331631
-
Potentiality of Soybean Proteomics in Untying the Mechanism of Flood and Drought Stress Tolerance.Proteomes. 2014 Mar 7;2(1):107-127. doi: 10.3390/proteomes2010107. Proteomes. 2014. PMID: 28250373 Free PMC article. Review.
-
Current views of drought research: experimental methods, adaptation mechanisms and regulatory strategies.Front Plant Sci. 2024 Apr 4;15:1371895. doi: 10.3389/fpls.2024.1371895. eCollection 2024. Front Plant Sci. 2024. PMID: 38638344 Free PMC article. Review.
Cited by
-
In search for drought-tolerant soybean: is the slow-wilting phenotype more than just a curiosity?J Exp Bot. 2020 Jan 7;71(2):457-460. doi: 10.1093/jxb/erz235. J Exp Bot. 2020. PMID: 31909814 Free PMC article.
-
Comparison of two different host plant genera responding to grapevine leafroll-associated virus 3 infection.Sci Rep. 2020 May 22;10(1):8505. doi: 10.1038/s41598-020-64972-8. Sci Rep. 2020. PMID: 32444786 Free PMC article.
-
Phosphorylation of Thr-225 and Ser-262 on ERD7 Promotes Age-Dependent and Stress-Induced Leaf Senescence through the Regulation of Hydrogen Peroxide Accumulation in Arabidopsis thaliana.Int J Mol Sci. 2024 Jan 22;25(2):1328. doi: 10.3390/ijms25021328. Int J Mol Sci. 2024. PMID: 38279327 Free PMC article.
-
Using the Knowledge of Post-transcriptional Regulations to Guide Gene Selections for Molecular Breeding in Soybean.Front Plant Sci. 2022 Mar 31;13:867731. doi: 10.3389/fpls.2022.867731. eCollection 2022. Front Plant Sci. 2022. PMID: 35432392 Free PMC article. Review.
-
Identification and Functional Analysis of a Protein Disulfide Isomerase (AtPDI1) in Arabidopsis thaliana.Front Plant Sci. 2018 Jul 19;9:913. doi: 10.3389/fpls.2018.00913. eCollection 2018. Front Plant Sci. 2018. PMID: 30073003 Free PMC article.
References
-
- Antolín M. C., Yoller J., Sánchez-Díaz M. (1995). Effects of temporary drought on nitrate-fed and nitrogen-fixing alfalfa plants. Plant Sci. 107 k159–165. 10.1016/0168-9452(95)04108-7 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

