Independent Evolution of Strychnine Recognition by Bitter Taste Receptor Subtypes
- PMID: 29552563
- PMCID: PMC5840161
- DOI: 10.3389/fmolb.2018.00009
Independent Evolution of Strychnine Recognition by Bitter Taste Receptor Subtypes
Erratum in
-
Corrigendum: Independent Evolution of Strychnine Recognition by Bitter Taste Receptor Subtypes.Front Mol Biosci. 2018 Sep 11;5:84. doi: 10.3389/fmolb.2018.00084. eCollection 2018. Front Mol Biosci. 2018. PMID: 30255025 Free PMC article.
Abstract
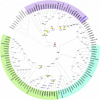
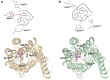
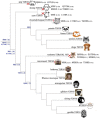
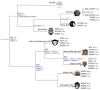
The 25 human bitter taste receptors (hT2Rs) recognize thousands of structurally and chemically diverse bitter substances. The binding modes of human bitter taste receptors hT2R10 and hT2R46, which are responsible for strychnine recognition, were previously established using site-directed mutagenesis, functional assays, and molecular modeling. Here we construct a phylogenetic tree and reconstruct ancestral sequences of the T2R10 and T2R46 clades. We next analyze the binding sites in view of experimental data to predict their ability to recognize strychnine. This analysis suggests that the common ancestor of hT2R10 and hT2R46 is unlikely to bind strychnine in the same mode as either of its two descendants. Estimation of relative divergence times shows that hT2R10 evolved earlier than hT2R46. Strychnine recognition was likely acquired first by the earliest common ancestor of the T2R10 clade before the separation of primates from other mammals, and was highly conserved within the clade. It was probably independently acquired by the common ancestor of T2R43-47 before the homo-ape speciation, lost in most T2Rs within this clade, but enhanced in the hT2R46 after humans diverged from the rest of primates. Our findings suggest hypothetical strychnine T2R receptors in several species, and serve as an experimental guide for further study. Improved understanding of how bitter taste receptors acquire the ability to be activated by particular ligands is valuable for the development of sensors for bitterness and for potential toxicity.
Keywords: ancestor functionality; ancestral reconstruction; bitter taste receptor; evolution; functional residues; homology modeling; ligand recognition; phylogenetics.
Figures
References
-
- Ballesteros J. A., Weinstein H. (1995). Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428. 10.1016/S1043-9471(05)80049-7 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

