Does the eclipse limit bacterial nucleoid complexity and cell width?
- PMID: 29552651
- PMCID: PMC5851910
- DOI: 10.1016/j.synbio.2017.11.004
Does the eclipse limit bacterial nucleoid complexity and cell width?
Abstract
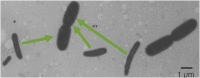
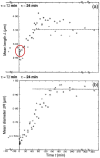
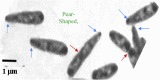
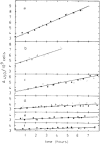
Cell size of bacteria M is related to 3 temporal parameters: chromosome replication time C, period from replication-termination to subsequent division D, and doubling time τ. Steady-state, bacillary cells grow exponentially by extending length L only, but their constant width W is larger at shorter τ's or longer C's, in proportion to the number of chromosome replication positions n (= C/τ), at least in Escherichia coli and Salmonella typhimurium. Extending C by thymine limitation of fast-growing thyA mutants result in continuous increase of M, associated with rising W, up to a limit before branching. A set of such puzzling observations is qualitatively consistent with the view that the actual cell mass (or volume) at the time of replication-initiation Mi (or Vi), usually relatively constant in growth at varying τ's, rises with time under thymine limitation of fast-growing, thymine-requiring E. coli strains. The hypothesis will be tested that presumes existence of a minimal distance lmin between successive moving replisomes, translated into the time needed for a replisome to reach lmin before a new replication-initiation at oriC is allowed, termed Eclipse E. Preliminary analysis of currently available data is inconsistent with a constant E under all conditions, hence other explanations and ways to test them are proposed in an attempt to elucidate these and other results. The complex hypothesis takes into account much of what is currently known about Bacterial Physiology: the relationships between cell dimensions, growth and cycle parameters, particularly nucleoid structure, replication and position, and the mode of peptidoglycan biosynthesis. Further experiments are mentioned that are necessary to test the discussed ideas and hypotheses.
Keywords: Bacterial cell cycle; Eclipse; Growth; Nucleoid complexity; Replication and division; Size and dimensions.
Figures


Similar articles
-
Does the Nucleoid Determine Cell Dimensions in Escherichia coli?Front Microbiol. 2019 Aug 6;10:1717. doi: 10.3389/fmicb.2019.01717. eCollection 2019. Front Microbiol. 2019. PMID: 31447799 Free PMC article.
-
Extending Validity of the Bacterial Cell Cycle Model through Thymine Limitation: A Personal View.Life (Basel). 2023 Mar 29;13(4):906. doi: 10.3390/life13040906. Life (Basel). 2023. PMID: 37109435 Free PMC article.
-
Changes of initiation mass and cell dimensions by the 'eclipse'.Mol Microbiol. 2007 Jan;63(1):15-21. doi: 10.1111/j.1365-2958.2006.05501.x. Epub 2006 Nov 27. Mol Microbiol. 2007. PMID: 17140410 Review.
-
Tight coupling of cell width to nucleoid structure in Escherichia coli.Biophys J. 2024 Feb 20;123(4):502-508. doi: 10.1016/j.bpj.2024.01.015. Epub 2024 Jan 19. Biophys J. 2024. PMID: 38243596 Free PMC article.
-
Instructive simulation of the bacterial cell division cycle.Microbiology (Reading). 2011 Jul;157(Pt 7):1876-1885. doi: 10.1099/mic.0.049403-0. Epub 2011 May 12. Microbiology (Reading). 2011. PMID: 21565934 Review.
Cited by
-
Hypothesis: bacteria live on the edge of phase transitions with a cell cycle regulated by a water-clock.Theory Biosci. 2024 Nov;143(4):253-277. doi: 10.1007/s12064-024-00427-2. Epub 2024 Nov 6. Theory Biosci. 2024. PMID: 39505803
-
Artificial modulation of cell width significantly affects the division time of Escherichia coli.Sci Rep. 2020 Oct 20;10(1):17847. doi: 10.1038/s41598-020-74778-3. Sci Rep. 2020. PMID: 33082450 Free PMC article.
-
Does the Nucleoid Determine Cell Dimensions in Escherichia coli?Front Microbiol. 2019 Aug 6;10:1717. doi: 10.3389/fmicb.2019.01717. eCollection 2019. Front Microbiol. 2019. PMID: 31447799 Free PMC article.
-
Open Questions about the Roles of DnaA, Related Proteins, and Hyperstructure Dynamics in the Cell Cycle.Life (Basel). 2023 Sep 10;13(9):1890. doi: 10.3390/life13091890. Life (Basel). 2023. PMID: 37763294 Free PMC article. Review.
-
Upconversion of Cellulosic Waste Into a Potential "Drop in Fuel" via Novel Catalyst Generated Using Desulfovibrio desulfuricans and a Consortium of Acidophilic Sulfidogens.Front Microbiol. 2019 May 10;10:970. doi: 10.3389/fmicb.2019.00970. eCollection 2019. Front Microbiol. 2019. PMID: 31134018 Free PMC article.
References
-
- Aarsman M.E.G., Piette A., Fraipont C., Vinkenvleugel T.M.F., Nguyen-Disteche M., den Blaauwen T. Maturation of Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55:1631–1645. - PubMed
-
- Amir A. Cell size regulation in bacteria. Phys Rev Lett. 2014;112:208102.
-
- Beacham I.R., Beacham K., Zaritsky A., Pritchard R.H. Intracellular thymidine triphosphate concentrations in wild type and in thymine requiring mutants of Escherichia coli 15 and K12. J Mol Biol. 1971;60:75–86. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

