Multidrug-Resistant Salmonella enterica 4,[5],12:i:- Sequence Type 34, New South Wales, Australia, 2016-2017
- PMID: 29553318
- PMCID: PMC5875280
- DOI: 10.3201/eid2404.171619
Multidrug-Resistant Salmonella enterica 4,[5],12:i:- Sequence Type 34, New South Wales, Australia, 2016-2017
Abstract
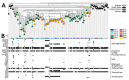
Multidrug- and colistin-resistant Salmonella enterica serotype 4,[5],12:i:- sequence type 34 is present in Europe and Asia. Using genomic surveillance, we determined that this sequence type is also endemic to Australia. Our findings highlight the public health benefits of genome sequencing-guided surveillance for monitoring the spread of multidrug-resistant mobile genes and isolates.
Keywords: Australia; Salmonella; antibiotic resistance; antimicrobial resistance; bacteria; genomics; molecular epidemiology; public health; salmonellosis; whole-genome sequencing.
Figures
References
-
- Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother. 2016;71:2300–5. 10.1093/jac/dkw093 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

